3769
Investigating the visual predicting model of metabolic biomarkers in biofluids for esophageal cancer detection using NMR-based metabolomics
Yan Lin1, Ting Ouyang1, Huanian Zhang1, Rongzhi Cai1, Peie Zheng1, Zhijie Fu1, Han Zhou1, and Renhua Wu1
1Radiology Department, Second Affiliated Hospital of Shantou University Medical College, Shantou City, China
1Radiology Department, Second Affiliated Hospital of Shantou University Medical College, Shantou City, China
Synopsis
Constructing an optimal metabolic model by combining biomarkers in biofluids may improve its non-invasive screening efficiency for esophageal cancer (EC). In this study, urine and serum specimens representing the healthy and EC individuals were examined using high-resolution 600 MHz 1H NMR technique. Furthermore, the paralleled patient-matched metabolites of EC tumor tissues and their adjacent non-cancerous tissues were investigated, which was used as references to determine biofluids metabolic biomarkers. The visual nomogram prediction model through a combination of creatine and glycine in both serum and urine was constructed using multiple regression analysis, which improves the diagnostic efficiency for EC.
Background
Identifying cancer-related biomarkers of esophageal cancer (EC) is essential for its early diagnosis and therapeutic intervention. Recently, we identified distinct nuclear magnetic resonance (1H-NMR)-based serum and urine metabolic signatures respectively, which were linked to the metabolic profiles of esophageal-cancerous tissues [1-3]. Our findings have highlighted the potential utility of NMR-based biofluids metabolomics fingerprinting as noninvasive predictors of earlier diagnosis in EC patients. However, the single biofluids metabolism can not fully present the metabolic characteristics of the whole body. Constructing an optimal metabolic model by combining biomarkers in serum and urine may achieve a comprehensive metabolic information of EC, therefore improving its non-invasive screening efficiency. In this study, urine and serum specimens representing the healthy and EC individuals were examined using high-resolution 600 MHz 1H NMR technique. Furthermore, the paralleled patient-matched metabolites of EC tumor tissues and their distant non-cancerous tissues were investigated, which was used as references to determine biofluids metabolic biomarkers. Pattern recognition was applied on NMR processed data to acquire the detailed metabolic information, and the predicting metabolic model through a combination of the biomarkers in serum and urine was constructed. Successful performance of this study could help to build up a new screening and diagnostic method, which is non-invasive, inexpensive, simple, and has high sensitivity and specificity for EC detection.Methods
Fifty EC patients with a scheduled esophageal resection participated in this study and provided esophageal cancer tissue (ECT) and distal noncancerous tissues (DNT, ~5 cm away from the tumor), together with the corresponding pre-operative serum and urine samples. Tissue samples were extracted with methanol/chloroform solution and the resulting supernatant was dried under vacuum for a minimum of 18 h. The lyophilized power of tissue, serum and urine samples were extracted with PBS/D20 buffer and a stock solution of TSP/D20 was added to each supernatant prior to analysis by 1H NMR spectroscopy. 1H-NMR spectra of esophageal tissues, serum and urine were detected by 600 MHz 1H-NMR spectrometer as previously described by us [1-6]. Generally, 1H-NMR spectra of esophageal tissue and urine were detected by using a NOESYPR1D pulse sequence, and serum spectra were acquired by a standard (1D) CarrPurcell-Meiboom-Gill (CPMG) pulse sequence. OPLS-DA model was used for pattern recognition analysis to identify potential biomarkers of EC tissues and biofluids. The Spearman correlation analysis was further employed to assess the associations of biomarker candidates between tissue and biofluids. The pathway analysis module of MetaboAnalyst software was used to analyze significant metabolic pathways. The nomogram and test curve were drawn by software R3.5.1. Double-tailed p values were used, and p<0.05 was considered to be statistically significant.Results
Representative 600 MHz 1H NMR spectra of tissue extracts obtained from ECT and DNT, together with serum and urine from EC and HC were shown in Fig 1. Good discrimination between cancer and their respective control was achieved by OPLS-DA scores plot generated from 1H NMR tissue spectra, serum and urine, respectively (Fig.2a). Model parameters of permutation analysis for different groups were as follows: ECT vs DNT: R2Y = 0.722, Q2 = 0.539; EC serum vs HC serum: R2Y = 0.986, Q2 = 0.978; EC urine vs HC urine: R2Y = 0.767, Q2 = 0.626, which indicated the good fit obtained by the model (Fig. 2b). Metabolic profiling associations across serum and urine potential biomarkers and tissue ones were analyzed, being plotted as correlation heat maps (Fig. 3), which showed that changes of serum isoleucine, leucine, lysine, glutamine, creatine, glycerol, myo-inositol and glucose in EC patients were correlated with changes of most metabolites in EC tissues (|r| >0.3, p<0.05). Alterations of creatine in EC urine were found to be associated with the changes of isoleucine, leucine, valine, arginine and lysine in EC tissues (|r| >0.3, p<0.05). The metabolic pathway analysis revealed that glycine, serine, and threonine metabolism was the most important, with pathway impact value of larger than 0.4 (p<0.05). Therefore, creatine and glycine in both serum and urine were selected as the best biomarkers, given that they were the major metabolites involved in glycine, serine, and threonine metabolism, and their changes in both serum and urine were related to changes of tissue metabolites in EC patients. Finally, logistic regression was performed to construct a visual nomogram diagnostic scoring model (Fig 4a), through a combination of creatine and glycine in both serum and urine. The diagnostic efficiency of this model was higher than any diagnostic model constructed by a single serum or urine metabolic markers, evidenced by a good prediction ability of 93% for EC detection (Fig 4b), and the prediction curve in the calibration graph and the standard curve fitted well (Fig 4c).Conclusion
The changes of urine and serum metabolism in esophageal cancer could reflect the metabolic disorder characteristics of the cancer tissues, highlighted the potential utility of NMR-based biofluids metabolomics fingerprinting as noninvasive predictors for EC detection. The visual nomogram prediction model based on creatine and glycine in both serum and urine can improve the diagnostic efficiency of EC. Further investigation is needed to validate these initial findings using larger samples and to establish the mechanism underlying progression of EC.Acknowledgements
This study was supported by grants from the National Natural Science Foundation of China (82071973, 82020108016) and Natural Science Foundation of Guangdong Province (2020A1515011022).References
1. Liang JH et al. World J Gastroenterol 2019;25:3218-30. 2. Ye W, et al. NMR Biomed 2021;34:e4505. 3. Y Lin, et al. ISMRM.26 (2020). 4.Yan Lin et al. Int J Cancer, 2019,145:1679-1689. 5. Yan Lin et al. Oncotarget, 2016, 7(20):29454-29464. 6. Y Lin, et al. ISMRM.25 (2017).Figures
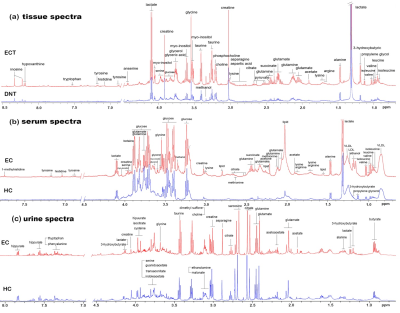
Fig 1. Representative 600
MHz 1H NMR spectra of tissue extracts obtained from ECT and DNT,
together with serum and urine obtained from EC and HC were shown in Figure 1. (a)
Representative NOESYPR1D 1H NMR spectra
of esophageal tissue extracts, (b) Representative
CMPG 1H NMR spectra of the serum, (c) Representative NOESYPR1D 1H NMR spectra
of urine.
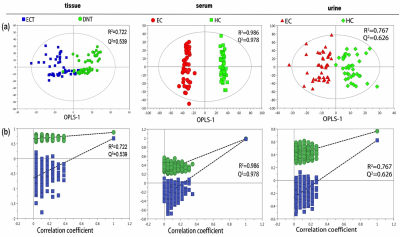
Fig
2. Pattern recognition analysis of 1H-NMR tissue, serum and urine
extract spectra between HC and EC. (a) OPLS-DA scores plot depicting the
difference between experimental groups; (b) Statistical validation of the corresponding
OPLS-DA model by permutation analysis (400 times).
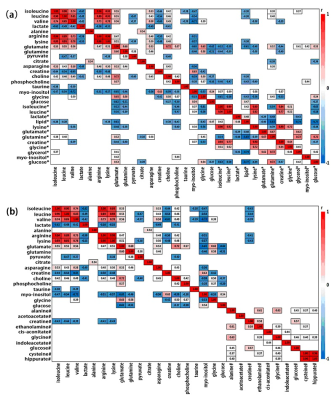
Figure 3. (a,b) Heat
map color-coded based on the strength of Spearman correlation
coefficients of metabolites identified as important in tumor vs serum and urine
discrimination. Cut-off values of |r| > 0.28 and p< 0.05 have been used (n=50).
The metabolites used are as given in table 1 (metabolites without label noted tissue
biomarkers; metabolites labeled with “*” noted serum
biomarkers; metabolites labeled with “#” noted urine biomarkers).
Red boxes indicate positive associations and blue boxes indicate negative
associations.
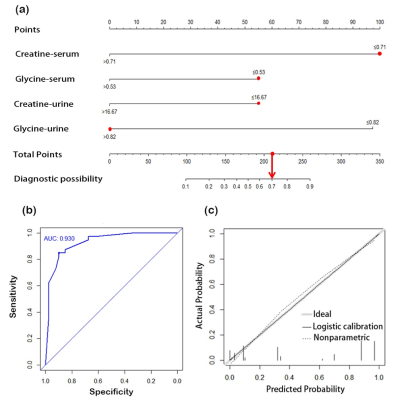
Fig 4. (a) Visual predictive nomogram
model based on serum and urine metabolic signature of EC patients. The No. 1
sample in modeling group has a total score of 210 points, indicating the risk
of esophageal cancer is 70%. (b) ROC curve of the nomogram model for predicting
esophageal cancer. (c) Calibration curve of the nomogram model for predicting
esophageal cancer.
DOI: https://doi.org/10.58530/2022/3769