3743
A Deep Survival Model based on MR Radiomics and EGFR status to Predict Overall Survival after Radiosurgery in Brain Metastases1Department of Biomedical Imaging and Radiological Sciences, National Yang Ming Chiao Tung University, Taipei, Taiwan, 2Department of Neurosurgery, Neurological Institute, Taipei Veteran General Hospital, Taipei, Taiwan, 3School of Medicine, National Yang Ming Chiao Tung University, Taipei, Taiwan, 4Brain Research Center, National Yang Ming Chiao Tung University, Taipei, Taiwan, 5Department of Radiology, Taipei Veteran General Hospital, Taipei, Taiwan, 6Department of Medical Imaging, Cheng-Hsin General Hospital, Taipei, Taiwan, 7Institute of Biophotonics, National Yang Ming Chiao Tung University, Taipei, Taiwan
Synopsis
About 30% of Non-small cell lung cancer (NSCLC) patients develop brain metastases (BMs) during the course of the disease. MR radiomics and EGFR mutation status were reported with the potential to predict the local tumor control of Gamma Knife stereotactic radiosurgery (GKRS). The prediction of overall survival after GKRS can further benefit the management of BM patients. We proposed a deep learning-based model using the clinical information, EGFR mutation status, and MR radiomic features to predict the overall survival after GKRS. We suggested that pre-GKRS MRI characteristics combined with gene and clinical information can improve the prediction of overall survival.
Background and Purpose
The brain metastasis (BM) of is a common complication of Non-small cell lung cancer (NSCLC). Median overall survival (OS) of NSCLC patients with BMs without additional therapy is about 1 month [1]. Due to the promising local tumor control rate, Gamma Knife radiosurgery (GKRS) has been one of the first-line treatments for BM [2]. MR radiomics and EGFR mutation states were indicated to show potential for predicting local tumor control in GKRS [3, 4]. However, the prediction of OS in BM patients remains challenging because multiple factors and complex situations may influence the patient outcome. In this study, we proposed a deep learning approach based on the EGFR status of primary tumor, pre-GKRS MRI radiomics and available clinical information to investigate the feasibility of OS prediction in BM patients after GKRS.Materials and Methods
We retrospectively collected data of 237 patients with BMs from NSCLC, and all the patients received GKRS treatment. Inclusion criteria included: 1) pathological diagnosis of NSCLC by lung biopsy or surgery; 2) diagnosis of brain metastases confirmed by MRI; 3) available EGFR mutation status information of primary NSCLC; and 4) available clinical and MRI follow-up after GKRS. The clinical characteristics of included BM patients are listed in Table 1. The EGFR status and relevant clinical features (Karnofsky performance status, KPS; existence of other metastasis besides BM; therapeutic effect of NSCLC; number of BMs; volume of BMs; additional treatment) were collected in this study. All the patients underwent the MR examinations before GKRS, including contrast-enhanced T1-weighted (T1c), T1-weighted (T1w), and T2-weighted (T2w) images. Several preprocessing steps were applied on the MRIs to improve the reliability of radiomics analysis. The image resolution was first adjusted by resampling voxel size to 1 x 1 x 1 mm3 for each MRI modality. The T1w and T2w images were then registered to T1c images using a six-parameter rigid body transformation and mutual information algorithm. The BM region of interest (ROI) was defined by radiation oncologists and reviewed by a neuro-radiologist for the GKRS treatment planning based on T1c and T2w images. To minimize the potential variability of radiomic features between multiple lesions within a single patient, only the radiomic features extracted from the largest BM were applied to the subsequent model training. Overall 1763 3D radiomic features, including histogram, geometric, texture and wavelet features were extracted from each BM ROI. The workflow of radiomic processing is displayed in Fig. 1. To identify key features for model training, univariate Cox proportional hazards models and Chi-squared tests were applied on the 70% of patients (training set) for selecting of radiomic and other (EGFR and clinical) features, respectively. A DeepSurv survival network (3 hidden layers, 8 nodes and 1000 epochs) was used to simulate the interactions between clinical or radiomic features and OS after GKRS [5]. The model performance was evaluated based on the remaining 30% of patients (test set). Predicted Kaplan-Meier (KM) survival curves were performed to visualize the DeepSurv OS modeling of patients. A log-rank test was applied to evaluate the statistical difference between the two KM curves. Time-dependent receiver operating characteristic (ROC) curves were used to evaluate the predictive effect of survival status in different time-point (3 months, 6 months, 12 months and 24 months).Results
A total of 6 radiomic features exhibited significant correlation (p<0.05) with OS after GKRS, including 3 histogram features from T1w images, and 3 histogram features from T1c images. The EGFR mutation status and other 3 clinical features were also selected by the feature selection method. Figure 2 shows the final selected features for model training. The simulated survival curves in the test set (Figure 3a) demonstrated the ability of the DeepSurv model to differentiate patients with longer or shorter survival time than the median OS (12.2 months). The average predicted survival curves between patients with longer and shorter OS also showed a statistically significant difference (p<0.05) using the log-rank test (Figure 3b). Figure 3c and 3d demonstrate the predicted survival curves of the representative BM patients (one shows a longer and another shows a shorter OS) with similar size and the same number of BMs. Time-dependent ROC curves at different survival time are showed in the Figure 4. The DeepSurv model demonstrated area under ROC curves (AUC) of 0.833, 0.831, 0.803 and 0.835, sensitivities of 0.789, 0.775, 0.714 and 0.857, and specificities of 0.786, 0.722, 0.806 and 0.807 in the OS of 3, 6, 12 and 24 months, respectively. Our results showed that the DeepSurv models were feasible in predicting OS of BM patients.Conclusions
In this study, we suggested that the deep learning model based on MR radiomics, EGFR status and clinical information could predict the overall survival of BM patients after GKRS. With our findings, more appropriate management or treatment strategies could be applied to treat BM patients.Acknowledgements
This work was supported by the Ministry of Science and Technology, Taiwan (MOST 109-2314-B-010-022-MY3) and Taipei Veterans General Hospital and University System of Taiwan (VGHUST110-G7-2-2).References
1. Kelly, K. and P.A. Bunn Jr, Is it time to reevaluate our approach to the treatment of brain metastases in patients with non-small cell lung cancer? Lung cancer, 1998. 20(2): p. 85-91.
2. Serizawa, T., et al., Gamma knife radiosurgery for metastatic brain tumors from lung cancer: a comparison between small cell and non—small cell carcinoma. Journal of neurosurgery, 2002. 97(Supplement 5): p. 484-488.
3. Liao, C.-Y., et al., Enhancement of Radiosurgical Treatment Outcome Prediction Using MRI Radiomics in Patients with Non-Small Cell Lung Cancer Brain Metastases. Cancers, 2021. 13(16): p. 4030.
4. Lee, C.-C., et al., Epidermal growth factor receptor mutations: association with favorable local tumor control following Gamma Knife radiosurgery in patients with non–small cell lung cancer and brain metastases. Journal of neurosurgery, 2019. 133(2): p. 313-320.
5. Katzman, J.L., et al., DeepSurv: personalized treatment recommender system using a Cox proportional hazards deep neural network. BMC medical research methodology, 2018. 18(1): p. 1-12.
Figures
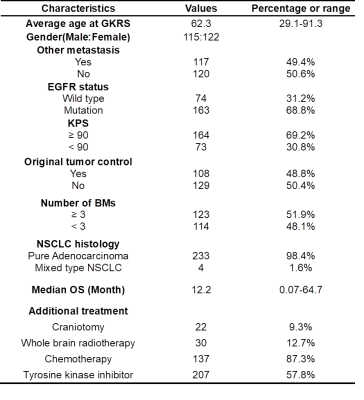
Table 1. Clinical characteristics of 237 patients
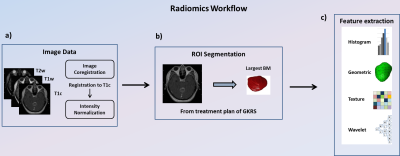
Figure 1. Image processing steps and radiomics flowchart
(a) Image acquisition of T1c, T1w and T2w images, registration of T1w and T2w to T1c images and intensity normalization. (b) ROI delineation for the treatment planning of GKRS by experienced neurosurgeons and radiologists on all MRI slices covering tumor regions. The largest BM of each patients was selected for the radiomic feature extraction. (c) Extraction of radiomic features from the tumor ROIs on the MRIs.
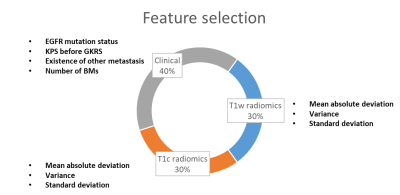
Figure 2. Final selected features for the prediction of overall survival of BM patients after GKRS.
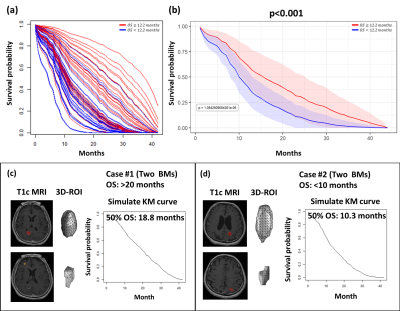
Figure 3. (a) The predicted KM survival curve of each patient in test set. (b) The average predicted KM curves of patients with survival longer and shorter than mean OS in the test set. The representative T1c images and predicted KM curves of patients with longer (c) and shorter (d) OS.
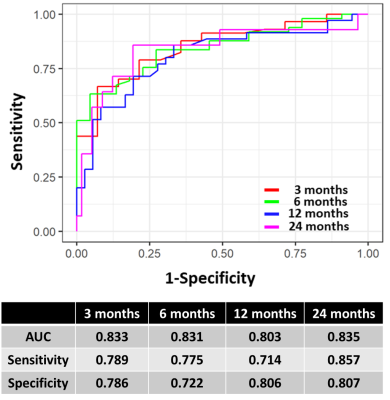
Figure 4. Time-dependent ROC curves of DeepSurv models at 3, 6, 12 and 24 months.