3742
A preliminary investigation of radiomics-based diffusion kurtosis imaging differences between glioblastoma and solitary brain metastasis
Eryuan Gao1, Guohua Zhao1, Huiting Zhang2, Ankang Gao1, Xiaoyue Ma1, Jie Bai1, Xu Yan2, Yusong Lin3, Guang Yang4, and Jingliang Cheng1
1department of magnetic resonance imaging, the First Affiliated Hospital of Zhengzhou University, Zhengzhou, China, 2MR Scientific Marketing, Siemens Healthcare, Shanghai, China, 3the School of Software, Zhengzhou University, Zhengzhou, China, 4Shanghai Key Laboratory of Magnetic Resonance, East China Normal University, Shanghai, China
1department of magnetic resonance imaging, the First Affiliated Hospital of Zhengzhou University, Zhengzhou, China, 2MR Scientific Marketing, Siemens Healthcare, Shanghai, China, 3the School of Software, Zhengzhou University, Zhengzhou, China, 4Shanghai Key Laboratory of Magnetic Resonance, East China Normal University, Shanghai, China
Synopsis
The differentiation of glioblastomas from solitary brain metastases by using routine magnetic resonance imaging (MRI) alone presents a challenge. As a representative of diffusion MRI technology, diffusion kurtosis imaging (DKI) can closely reflect the real situation of water molecule movement in tumor tissues. We speculate that DKI radiomics is superior to routine MRI radiomics in distinguishing GBMs from SBMs. We developed a series of radiomics models of DKI parameter maps and routine MRI to compare their performance. Finally, several independent parameters models perform well in distinguishing two tumors. The combined DKI radiomics model obtained the best performance.
Introduction
Glioblastomas (GBMs) and solitary brain metastases (SBMs) are types of brain tumor that commonly occur in adults; their incidence is approximately half of all cases [1]. The differentiation of GBM from SBMs by using routine magnetic resonance imaging (MRI) alone is a challenge given that both tumors present with imaging patterns consisting of peritumoral hyperintensities with similar intratumoral textures. Radiomics has shown great potential in the computer-aided diagnosis of tumors. Routine MRI radiomics analysis has satisfactory classification capability in the discrimination of GBMs and SBMs [2]. In addition, advanced imaging modalities, such as diffusion kurtosis imaging (DKI), can effectively distinguish the two tumors. As a representative of diffusion MRI technology, DKI can closely reflect the real situation of water molecule movement in tumor tissues [3]. Based on the above point of view, we speculate that DKI radiomics is superior to routine MRI radiomics in distinguishing GBMs from SBMs. Therefore, this research aimed to explore the performance of DKI radiomics analysis in distinguishing the two types of tumors.Materials and Methods
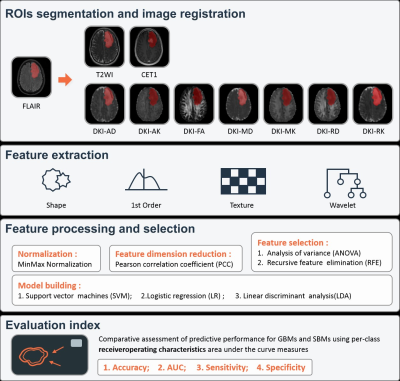
The institutional review board approved this prospective study, and informed consent was obtained from every patient. We recruited GBM and SBM patients between November 2015 and April 2021. The inclusion criteria were as follows: (1) pathologically confirmed as having GBMs and (2) pathologically confirmed as having SBMs or follow-up confirmation of SBMs; (3) presence of T2WI, FLAIR, CE-T1WI, and diffusion weighted imaging (DWI) sequences. The exclusion criteria were as follows: (1) lack of necessary MRI scans and (2) MRI scans with severe motion or susceptibility artifacts. All patients underwent DWI and routine MRI examinations on a 3T MR scanner (MAGNETOM Prisma; Siemens Healthcare, Erlangen, Germany) with a 64-channel head–neck coil. Finally, 101 patients, including 50 GBMs and 51 SBMs, were included. We randomly selected 81 cases as the training cohort, and the other 20 cases as the independent testing cohort. DWI data were acquired using 6b values (0, 500, 1000, 1500, 2000, and 2500 s/mm2), and every non-zero b value was performed at 30 encoding directions. The DKI parameters were calculated from the DWI data by an in-house-developed post-processing software, NeuDilab, based on DIPY (http://nipy.org/dipy). The DKI parameters included axial, radial, and mean diffusivities (AD, RD and MD, respectively); axial, radial, and mean kurtosis (AK, RK, and MK, respectively); fractional anisotropy (FA). Using the ITK-SNAP (http://www.itksnap.org) software, we opened the axial FLAIR image and manually drew the outline of the maximum abnormal signal area on each slice showing the tumor, and the results were saved as the segmented regions of interest (ROIs). Then, routine MRI images and all DKI parameter maps were spatially registered to FLAIR images. Using the FAE software, we extracted 851 radiomics features from each case, including 18 first-order statistical features, 14 shape-based, and 75 texture features of original and wavelet transformed images [4]. Two feature selection methods and three classifiers were utilized to construct radiomics prediction models of routine MRI and individual DKI parameter maps. In addition, a combined routine MRI model and a combined DKI model were constructed. Fivefold cross-validation was applied to demonstrate the model performance, which was evaluated using the receiver operating characteristic (ROC) curve, accuracy, area under the ROC curve (AUC), sensitivity, and specificity on the testing cohort.Results
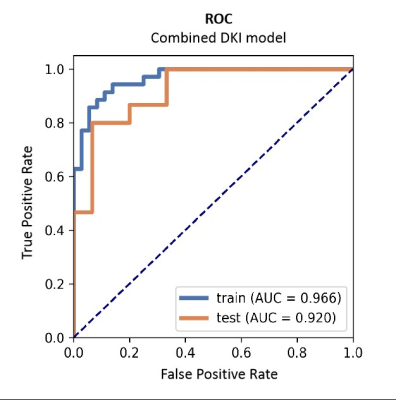
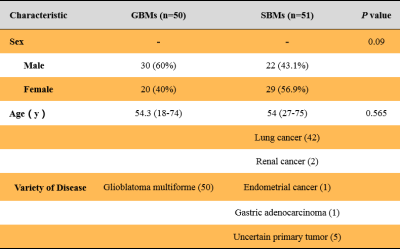
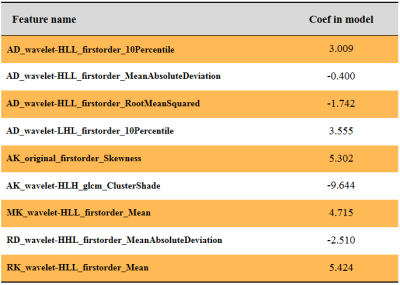
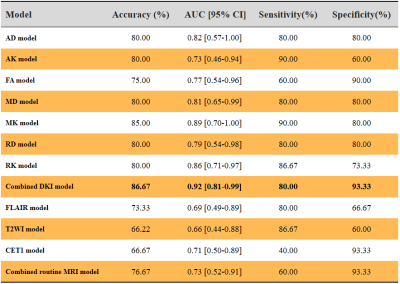
Table 1 summarizes the detailed clinical characteristics. No significant difference was found between two tumors regardless of sex (P = 0.09) and age (P = 0.565). Figure 2 shows the best performance obtained with the combined DKI model. The model used analysis of variance for feature selection and selected nine key features to serve as the radiomics signatures (Table 2). The radiomics signatures were used to build a predictive model by the logistic regression classifier. Table 3 shows the performance of each parameter and the two combined models of DKI parameter and routine MRI. The combined routine MRI model achieved accuracy 76.67%, AUC of 0.73, and sensitivity of 60.00%, whereas the combined DKI model showed the best performance (accuracy: 86.67%, AUC: 0.92, and sensitivity: 93.33%) for distinguishing between GBMs from SBMs in the independent testing cohort.Discussion
Previous studies have shown that AK, RK, RD, MD, and MK exhibit significant differences between GBMs and SBMs [2-3]. Our results were consistent with these findings. Independent models of these parameters performed well in differentiating the two tumors. Investigators have shown that the peritumoral FA was not significantly different when comparing GBMs with SBMs [3]. Here, the ROIs included peritumoral ones, and the differential capability of FA was inferior to that of other DKI parameters. In general, the combined DKI model showed excellent discrimination capability. Besides, the combined routine MRI model had a dissatisfactory performance in distinguishing the two tumors. GBMs and SBMs reflect a high degree of tissue complexity, including increased cell density and high nuclear atypia [5]. Routine MRI has a very limited capability to distinguish between the two tumors. DKI parameters were more sensitive to changes in the micro-structure of tissues, which can be easily extracted and used as radiomics features, than routine MRI.Acknowledgements
No acknowledgement found.References
[1] Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 2016;131(6):803-20. [2] Mao J, Zeng W, Zhang Q, et al. Differentiation between high-grade gliomas and solitary brain metastases: a comparison of five diffusion-weighted MRI models. BMC Med Imaging. 2020;20(1):124. [3] Tan Y, Wang XC, Zhang H, et al. Differentiation of high-grade-astrocytomas from solitary-brain-metastases: Comparing diffusion kurtosis imaging and diffusion tensor imaging. Eur J Radiol. 2015;84(12):2618-24. [4] Song Y, Zhang J, Zhang YD, et al. FeAture Explorer (FAE): A tool for developing and comparing radiomics models. PLoS One. 2020;15(8):e0237587. [5] Lee EJ, Ahn KJ, Lee EK, et al. Potential role of advanced MRI techniques for the peritumoural region in differentiating glioblastoma multiforme and solitary metastatic lesions. Clin Radiol. 2013;68(12):e689-97.Figures
DOI: https://doi.org/10.58530/2022/3742