3703
An automatic pipeline combining deep learning and radiomics to predict placenta accrete spectrum disorders from T2W MR images1Shanghai key lab of magnetic resonance, East China Normal University, Shanghai, China, 2Department of Radiology, Obstetrics and Gynecology Hospital of Fudan University, Shanghai, China, 3Department of Obstetrics, Obstetrics and Gynecology Hospital of Fudan University, Shanghai, China
Synopsis
Placenta accrete spectrum (PAS) disorders may lead to common complication like catastrophic perinatal hemorrhage. T2W is the most useful MRI sequence for identification of PAS disorders, but the diagnosis of PAS is often difficult and highly subjective. To overcome this problem, we automatically segmented the placental regions by nnU-Net and used radiomics features extracted from the segmented region to build a radiomics-clinical model for identification of placenta invasion. 512 pregnant women were enrolled in this study. Our segmentation model achieved a mean Dice coefficient of 0.890 and the classification model achieved an AUC of 0.849 on the independent validation cohort.
Introduction
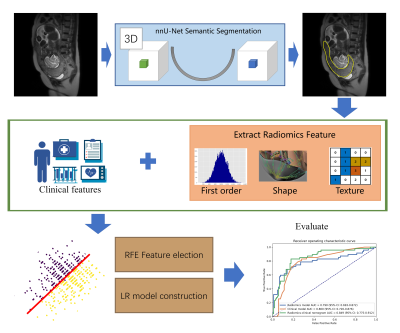
Placenta accrete spectrum (PAS) disorders is a general term used to describe abnormal trophoblastic invasion into myometrium of the uterine wall. PAS occur mostly in patients with placenta previa after a prior cesarean section delivery with an increasing number of cesarean deliveries1, which is prone to a variety of complications, such as hemorrhage during or after delivery, disseminated intravascular coagulation, renal failure, venous thrombosis, etc., which may lead to maternal or fetal death in severe cases2,3. The placental heterogeneity on T2W images and irregular thick black bands within the placenta are well-recognized signs of placental infiltration in MR images4,5. However, identification of such signs requires expertise of experienced radiologists and can be difficult for junior radiologists5,6. Radiomics models require region of interest (ROI) be outlined first. Manual outlining of ROIs increases the workload of the radiologist and often suffers from inter-reader variance. Thus, we proposed an approach to automatically segment the placental region on T2W images by nnU-Net and build a radiomics-clinical model based on the automatically segmented ROIs to identify PAS disorders.Methods
A total of 512 suspicious PAS disorders pregnant women T2W MR images were collected from a 1.5-T MR system (Magnetom Avanto, Siemens) between January 2015 and December 2018 at Obstetrics and Gynecology Hospital of Fudan University. According to the gold standard of clinical/pathological outcome of pregnancy, 157 cases of PAS were diagnosed. The region of placenta was labeled by a radiologist with 10 years of experience. We randomly split the dataset into a training cohort (110 PAS disorders / 247 normal) and a validation cohort (47 PAS disorders /108 normal).A flowchart of this study was shown in Figure 1. A semantic segmentation model was trained with nnU-Net7 to segment the placental region to be used for radiomics feature extraction. The nnU-Net can automatically adapt to arbitrary datasets and take full advantage of the characteristics of the dataset to outperform many U-Net based models, and has been used successfully in 3D biomedical image segmentation. We used the combination of cross entropy loss and dice loss as the loss function. The stochastic gradient descent (SGD) algorithm was used as optimizer with an initial learning rate of 0.01, a momentum of 0.9 and weight decay was 0.005.
Then we used an open-source software FeatureExplorer8 for feature extraction and model building. In the backend, FeatureExplorer uses PyRadiomics (version 3.0)9 for feature extraction, and scikit-learn for machine learning model building. It can automatically try combinations of different algorithms find the best classification model according to its performance in a n-fold cross-validation on the training cohort. From each case, 107 radiomics features were extracted, including the shape, first order and texture features. The combination of Pearson correlation coefficient (PCC) for dimension reduction, recursive feature elimination (RFE) for feature selection, and logistic regression (LR) classifier was found to yield the best radiomics model.
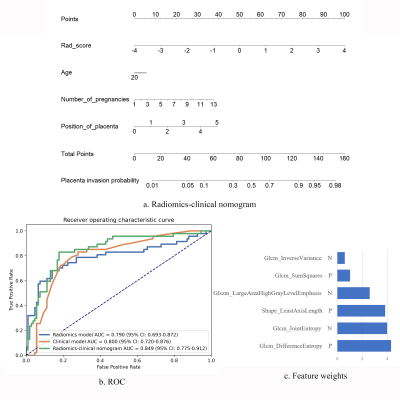
A clinical LR model based on clinical characteristics, such as placental location, maternal age, and gravida and para, was also built. Further, a radiomics-clinical nomogram was constructed with the radiomics score calculated with the radiomics model and the clinical variables to predict the risk of PAS disorders.
Results
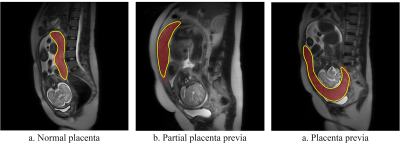
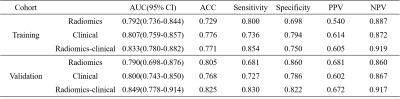
The trained nnU-Net model got a mean Dice coefficient of 0.890 in the validation cohort. As shown in Figure 2, the segmentation was in good agreement with the radiologists' label.The final radiomics model included 6 features (Figure 3c) and achieved an AUC of 0.790 (95% CI, 0.698–0.876). The clinical model achieved an AUC of 0.800 (95% CI, 0.743–0.850). The combined radiomics-clinical nomogram (Figure 3a) achieved a higher AUC of 0.849 (95% CI, 0.778–0.914). The statistics analysis for clinical/radiomics/radiomics-clinical nomogram in the two cohorts were listed in Table 1 and the ROC curves were showed in Figure 3b.
Discussion
In this study, a trained nnU-Net model was utilized to automatically segment placenta region on T2W MR images. Compared with previous radiomics study, automatic segmentation of ROI could reduce subjectivity, the workload of radiologist and the negative impact of inter-reader variance on the prediction. The results showed that the radiomics-clinical model achieved a satisfying performance in PAS disorders identification. The radiomics texture features reflect the intra-placenta heterogeneity, which may indicate the physiological change such as fibrin deposition. Clinical features including placental location, maternal age, and gravida and para played an equally important role. It was confirmed that PAS disorders are influenced by a combination of several factors.There are some limitations of current research. This retrospective study in a single center is lack of effective external validation and some enrolled patients may have other complications, which may affect the analysis.
Conclusion
In conclusion, a radiomics-clinical model using features extracted from automatically segmented placental regions was established and can be used to accurately predict risk of PAS disorders during pregnancy, which may effectively improve the accuracy of PAS disorders diagnosis in primary hospitals, provide timely and reasonable referral, and reduce the maternal mortality rate.Acknowledgements
This project is supported by National Natural Science Foundation of China (61731009).References
1. Thurn L, Lindqvist PG, Jakobsson M, et al. Abnormally invasive placenta-prevalence, risk factors and antenatal suspicion: results from a large population-based pregnancy cohort study in the Nordic countries. BJOG : an international journal of obstetrics and gynaecology 2016;123:1348-55
2. Fitzpatrick, K. E. et al. Incidence and Risk Factors for Placenta Accreta/Increta/Percreta in the UK: A National Case-Control Study. Plos One. 2012;7(12).
3. Oyelese, Y. & Smulian, J. C. Placenta previa, placenta accreta, and vasa previa. Obstetrics and Gynecology. 2006;107(4):927-941.
4. Lax, A., Prince, M. R., Mennitt, K. W., Schwebach, J. R. & Budorick, N. E. The value of specific MRI features in the evaluation of suspected placental invasion. Magnetic Resonance Imaging. 2007;25(1):87-93.
5. Shao, Q. et al. Deep learning and radiomics analysis for prediction of placenta invasion based on T2WI. Mathematical Biosciences and Engineering. 2021;18(5):6198-6215.
6. Alamo, L. et al. Detection of suspected placental invasion by MRI: Do the results depend on observer' experience? European Journal of Radiology. 2013;82(2):E51-E57.
7. Isensee, F., Jaeger, P. F., Kohl, S. A. A., Petersen, J. & Maier-Hein, K. H. nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nature Methods. 2021;18(2):203.
8. Song, Y. et al. FeAture Explorer (FAE): A tool for developing and comparing radiomics models. Plos One. 2020;15(8).
9. Van Griethuysen, J. J. M., Fedorov, A., Parmar, C., Hosny, A., Aucoin, N., Narayan, V., Beets-Tan, R. G. H., Fillon-Robin, J. C., Pieper, S., Aerts, H. J. W. L. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Research. 2017;77(21):e104–e107.
Figures