3697
Kidney segmentation in MR images using CT-trained ResUNet and transfer learning1Vascular and Physiologic Imaging Research (VPIR) Lab, School of Biomedical Engineering, ShanghaiTech University, Shanghai, China, 2X-ray Systems Lab, School of Biomedical Engineering, ShanghaiTech University, Shanghai, China, 3SAIIP, School of Computer Science and Technology, Northwestern Polytechnical University, Xi'an, China
Synopsis
Kidney segmentation is often necessary for analyzing renal MRI data. Deep learning approach shows much promise, but typically requires large numbers of images for model training. In this study, we explored the feasibility of segmenting MRI images using ResUNet pre-trained with CT images and fine-tuning with transfer learning. The fine-tuning step used 60 MRI images (from 5 subjects) only. The trained model performed excellently in segmenting an independent set of MRI images, with DICE similarity of 0.94 and volume error of 13%±9%. This study demonstrates the power of transfer learning in utilizing images of a different modality in kidney segmentation.
Introduction
MRI has been widely used for imaging kidneys, for high-resolution structure characterization and for assessment of various physiologic aspects 1-3. In analyzing the MRI images, kidney segmentation is often a necessary pre-processing step. Manual delineation by a human operator is usually time consuming and sometimes involves large inter-observer variability. Deep learning with neural networks has shown much promise in segmenting kidney MRI images 4. However, training such a deep network requires a large number (e.g. tens of thousands) of images that are similar to the images to be segmented. Recently the idea of transfer learning was proposed, making it possible for the model to be pre-trained with images of a different type. In this study, we used a large set of CT images to train a ResUNet model, fine-tuned it with a small set of MRI images, and tested the model’s performance in segmenting a different set of kidney MRI images.Methods
For kidney segmentation, we chose a Deep-Residual U-Net (ResUNet) model 5,6 (Fig. 1), which combines the strength of deep residual learning and of U-Net. U-Net is one of the most powerful deep learning architectures for medical image segmentation 5,7. The residual block solves the degradation problem in the encoders of deep U-Net, so improves both the channel inter-dependencies and computational efficiency. For pre-training the ResUNet model, we used about 4000 CT kidney images (200 cases, each with 20 coronal slices) from the KiTS19 challenge. To fine-tune the model with transfer learning, we used BOLD images acquired from 5 healthy subjects; each subject’s dataset contained 12 images with different echo times (4.3-42.7 ms). The MR data were acquired at 3T (Tim Trio; Siemens Medical Solutions).The model training was implemented on a PC workstation equipped with a single NVIDIA GeForce RTX 3070 GPU and Inter(R) Xeon(R) Gold 5220R CPU @2.20GHz x 48, and in an environment with Ubuntu 20.04.3 LTS OS and the Cuda version 11.2. The pre-training step used 4000 CT images, with a batch size of 1 and Adam optimizer. The learning rate was set to 1e-4 initially and decayed it with the ReduceLROnPlateau method. In the fine-tune step, we started the learning from the pre-trained parameter values, set the learning rate to 1e-4, then trained the model with the provided 60 MRI images. To evaluate the performance of the trained model, we used it to segment an independent set of 7 BOLD images, and compared it against manual segmentation by a human operator. For comparison, we also applied the pre-trained model (without transfer learning) and the model trained with the 60 MRI images in segmenting the same 7 BOLD images.
Results
Pre-training of the ResUNet model with CT data was completed with an accuracy of 0.92, and transfer learning to MRI images had an accuracy of 0.94. Fig. 2 shows representative examples of segmenting with a ResUNet trained with 60 MRI images only (no pre-training with CT images) and with the proposed method. Sixty MRI images were far few for training the ResUNet, so that its segmentation captured only part of the kidneys (Fig. 2b). Pre-training with large number of CT images tremendously improved the segmentation, even though the shown case (Fig. 2c) had some susceptibility artifact in the lower lobe of the left kidney. In Fig.2, we did not show any segmentation example for the pre-trained ResUNet (no fine-tuning), because such model did not output any mask for MRI images. When compared to the manual segmentation across the 7 BOLD images, the proposed method had segmentation error of 13%±9%.Discussion
In reality, it is often challenging to accumulate thousands of images for training a deep neural network, and to label the images is also time consuming. This study reveals the feasibility of using a large set of CT images (with manually segmented masks) to pre-train a ResUNet, and then fine-tuning the model with a rather small set of MRI images. The performance of the model is satisfactory with segmentation error on the order of 10%. It was also noted that even though the model was pre-trained with images of a totally different modality, its final performance was not compromised. For example, susceptibility artifact is very common for renal BOLD, displayed as dark regions eroding into kidneys (Fig. 2C). However, such regions were properly segmented, suggesting the robustness of the model. Similarly, the model handled the segmentation of the kidney with its other surrounding organs nicely, even though the contrast in MRI was quite different from that in CT images. As a minor problem, for some cases the proposed method mistakenly masked some muscle areas (upper-left corner in Fig. 2C). This problem can be easily corrected by a simple image cropping, or more elegantly by utilizing data augmentation to improve the model.Conclusion
In conclusion, a ResUNet pre-trained with CT images and fine–tuned with a small set of MRI images is capable of segmenting kidney MRI images excellently.Acknowledgements
No acknowledgement found.References
1. Zollner FG, Kocinski M, Hansen L, et al. Kidney Segmentation in Renal Magnetic Resonance Imaging - Current Status and Prospects. IEEE Access. 2021;9:71577-71605. doi:10.1109/access.2021.3078430
2. Zöllner FG, Svarstad E, Munthe-Kaas AZ, Schad LR, Lundervold A, Rørvik J. Assessment of kidney volumes from MRI: acquisition and segmentation techniques. American Journal of Roentgenology. 2012;199(5):1060-1069.
3. Torres HR, Queiros S, Morais P, Oliveira B, Fonseca JC, Vilaca JL. Kidney segmentation in ultrasound, magnetic resonance and computed tomography images: A systematic review. Computer methods and programs in biomedicine. 2018;157:49-67.
4. Caroli A, Schneider M, Friedli I, et al. Diffusion-weighted magnetic resonance imaging to assess diffuse renal pathology: a systematic review and statement paper. Nephrology Dialysis Transplantation. 2018;33(suppl_2):ii29-ii40.
5. Jha D, Smedsrud PH, Riegler MA, et al. Resunet++: An advanced architecture for medical image segmentation. IEEE; 2019:225-2255.
6. Ronneberger O, Fischer P, Brox T. U-net: Convolutional networks for biomedical image segmentation. Springer; 2015:234-241.
7. Zhang Z, Liu Q, Wang Y. Road extraction by deep residual u-net. IEEE Geoscience and Remote Sensing Letters. 2018;15(5):749-753.
Figures
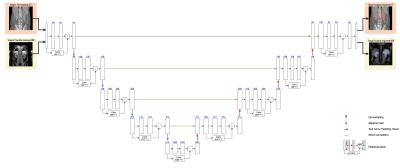
Fig. 1: The ResUNet model architecture contains an encoder pathway (the left side) and a decoder pathway (right side), each including four blocks of down-sampling or up-sampling coupled with residual learning. Pre-training was done with CT data (top row of images), followed by fine-tuning all the layers with a small set of MRI images (second row of images).
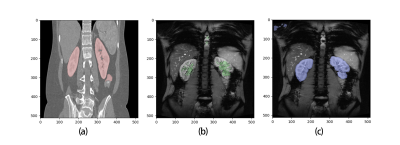
Fig.2. Representative of segmentation results. a) CT image segmentation by the pre-trained model; b) MRI image segmentation by a ResUNet model trained with 60 MRI images: the result was bad due to the small number of training images; c) MRI image segmentation by the proposed method, i.e. ResUNet pre-trained with CT images and then fine-tuned with 60 MRI images.