3686
Radiomics Analysis of the Prostate Cancer Habitats in Diagnosing Extracapsular Extension on Multi-Parametric MRI1Shanghai Key Laboratory of Magnetic Resonance, East China Normal University, Shanghai, China, 2Department of Radiology, the First Affiliated Hospital with Nanjing Medical University, Nanjing, China, 3Department of Radiology, the First Affiliated Hospital with Soochow University, Soochow, China, 4MR Scientific Marketing, Siemens Healthineers Ltd, Shanghai, China
Synopsis
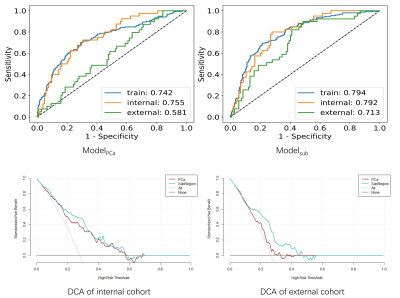
We extracted radiomics features from subregions of the area where the lesion was close to the capsular boundary to diagnose the extracapsular extension (ECE). The model was trained with 574 cases, using 5-fold cross-validation and evaluated with an independent internal test cohort of 144 cases and an external test cohort of 146 cases. The model built with subregion features achieved AUC (area under receiver operating characteristic curve) values of 0.792 and 0.713 on the internal and external test cohort, respectively, outperforming the model built with whole lesion features whose internal and external test AUCs are 0.724 and 0.581, respectively.
INTRODUCTION
The presence of extracapsular extension (ECE) of prostate cancer (PCa) has an influence on the strategy of surgery. Studies have shown that multi-parametric magnetic resonance imaging (MRI) can be used to effectively detect the ECE1-3. However, the diagnosis still requires the expertise of the physician and suffers from low specificity4, 5.Radiomics6 was widely used in analyzing medical images by extracting quantitative features from the region of interest (ROI). Previous studies showed that radiomics features extracted from lesions could help diagnose the ECE7, 8. However, the ECE diagnosis should depend on not only the lesion features, but also its relationship with surrounding tissues9.
In this study, inspired by Sherif Mehralivand’s work9, we focused on the region of the lesion close to the boundary of the prostate gland. We split the region into three habitats: lesion, prostate gland, and the surrounding tissues, and extracted features from each habitat. We compared proposed subregion model (Modelsub) with the whole lesion model (ModelPCa) built with features of the whole lesion.
METHODS
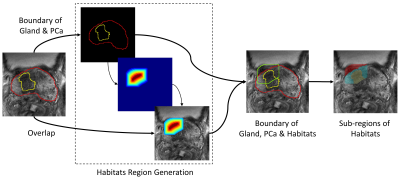
The data in our retrospective study was collected from the Jiangsu Province Hospital (JSPH) and the First Affiliated Hospital of Soochow University (SUH), and the study was approved by both institutional ethics committees. 718 cases (191 cases with ECE) from JSPH were obtained on two 3T scanners (Skyra, Siemens Healthcare and MR770, United Imaging Healthcare). Additional 146 (33 positives) cases scanned on Siemens Healthcare 3T Skyra were from SUH. Protocols suggested by PIRADS v2.1 were used to get T2-weighted images (T2WI) and apparent diffusion coefficient (ADC) maps. Data from JSPH was randomly split into a training cohort (574, 151 positives) and an internal validation cohort (144, 40 positives), while SUH data was used for external vallidation.Two experienced genitourinary radiologists. labeled the ROIs of the PCa and the prostate gland on T2WI manually. Based on these two ROIs, we generated a candidate region with a high probability of occurring ECE, which includes lesion, prostate gland, and the surrounding tissues10, shown in Figure 1. Then three sub-regions were segmented based on this candidate region and labeled ROIs. ADC maps were aligned to T2WI with Elastix. All images were resampled to an inner-slice resolution of 0.5×0.5mm2. Shape-based and histogram-based features were extracted using sub-regions, T2WI images, and ADC maps.
The radiomics workflow was illustrated in Figure 2. We used the synthetic minority oversampling technique (SMOTE) to balance the training data set. Combinations of three different normalizations, three feature selection methods, and four classifies were iterated to find the best model, which model was selected based on 5-fold cross validation. Then we compared our Modelsub to the ModelPCa by receiver operating characteristic (ROC) curve, decision curve analysis (DCA) and confusion matrix. Delong test was used to compare the models’ performance and 0.05 was used for significance. All above process was implemented with FeAture Explorer (FAE, V 0.4.3)11.
RESULTS
For ModelPCa, the best model is logistical regression (LR) with 12 features, which achieved an AUC of 0.742/0.755/0.581 on training/internal validation/external validation. Sensitivities on all three cohorts were over 0.7, indicating that the model could identify patients with ECE. For Modelsub, an LR model using 14 features achieved AUCs of 0.794/0.792/0.713 on training/internal validation/external validation cohort. Modelsub was significantly better than ModelPCa (p < 0.05). The clinical statistics, ROC and DCA curves of the two models was shown in Figure 3 and Figure 4, respectively.DISCUSSION and CONCLUSION
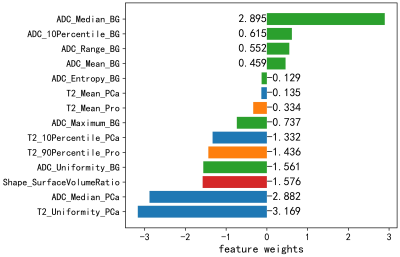
We built a radiomics model using features extracted from the sub-regions from where PCa is close to gland boundry. Modelsub achieved test AUCs of 0.792 and 0.714 on the internal and external test cohort, much higher than those of 0.755 and 0.581 with ModelPCa. This performance improvement indicates that the sub-region analysis of the lesion habitats can identify ECE better. As shown in Figure 5, the Modelsub kept 14 features, including 7 surrounding region features, 4 PCa features, 2 prostate features, and 1 shape feature, which denotes that our sub-region analysis of PCa habitats could utilize all information of different regions for more accurate ECE diagnosis.It is understandable that volume surface ratio feature had a significant contribution to the prediction, because if the lesion breaks through the gland, it will surely make the shape of generated attention ROI more complex, wiith an increased surface. More than half of the features retained in the model were histogram features from different subregions on ADC map, which maybe attributed to the fact that ADC maps exhibited high contrast between differen tissues and lesions.
There are some limitations in our study. The habitats region was generated by the manually labeled prostate gland and PCa, which might be replaced by the deep learning segmentation and detection. Additional MRI sequences may provide more information for ECE diagnosis and should be explored in the future.
In conclusion, radiomics features of PCa habitats could be used to help ECE diagnosis on MRI.
Acknowledgements
No acknowledgement found.References
1. Cerantola Y, Valerio M, Kawkabani Marchini A, Meuwly J-Y, Jichlinski P. Can 3T multiparametric magnetic resonance imaging accurately detect prostate cancer extracapsular extension? Canadian Urological Association journal = Journal de l'Association des urologues du Canada. 2013;7(11-12):E699-E703.
2. Feng TS, Sharif-Afshar AR, Smith SC, et al. Multiparametric magnetic resonance imaging localizes established extracapsular extension of prostate cancer. Urologic Oncology: Seminars and Original Investigations. 2015;33(3):109.e115-109.e122.
3. Roethke MC, Lichy MP, Kniess M, et al. Accuracy of preoperative endorectal MRI in predicting extracapsular extension and influence on neurovascular bundle sparing in radical prostatectomy. World Journal of Urology. 2013;31(5):1111-1116.
4. Freifeld Y, Diaz de Leon A, Xi Y, et al. Diagnostic Performance of Prospectively Assigned Likert Scale Scores to Determine Extraprostatic Extension and Seminal Vesicle Invasion With Multiparametric MRI of the Prostate. American Journal of Roentgenology. 2018;212(3):576-581.
5. Ma S, Xie H, Wang H, et al. Preoperative Prediction of Extracapsular Extension: Radiomics Signature Based on Magnetic Resonance Imaging to Stage Prostate Cancer. Molecular Imaging and Biology. 2020;22(3):711-721.
6. Lambin P, Rios-Velazquez E, Leijenaar R, et al. Radiomics: Extracting more information from medical images using advanced feature analysis. European Journal of Cancer. 2012;48(4):441-446.
7. Xu L, Zhang G, Zhao L, et al. Radiomics Based on Multiparametric Magnetic Resonance Imaging to Predict Extraprostatic Extension of Prostate Cancer2020;10(940).
8. He D, Wang X, Fu C, et al. MRI-based radiomics models to assess prostate cancer, extracapsular extension and positive surgical margins. Cancer Imaging. 2021;21(1):46.
9. Mehralivand S, Shih JH, Harmon S, et al. A Grading System for the Assessment of Risk of Extraprostatic Extension of Prostate Cancer at Multiparametric MRI. Radiology. 2019;290(3):709-719.
10. Hou Y, Zhang Y-H, Bao J, et al. Artificial intelligence is a promising prospect for the detection of prostate cancer extracapsular extension with mpMRI: a two-center comparative study. European Journal of Nuclear Medicine and Molecular Imaging. 2021;48(12):3805-3816.
11. Song Y, Zhang J, Zhang Y-d, et al. FeAture Explorer (FAE): A tool for developing and comparing radiomics models. PLOS ONE. 2020;15(8):e0237587.
Figures