3677
Automatic segmentation of prostate gland on multiparametric MR images using deep learning convolutional neural network: a multi-center study1Peking Union Medical College Hospital, Beijing, China, 2Deepwise AI Lab, Beijing, China
Synopsis
Accurate prostate segmentation on MR images plays an important role in the management of prostate diseases. Recently proposed deep learning architecture has been successfully applied for medical image segmentation to overcome the shortcomings of manual segmentation. Our study proposed a 3D UNet model for automatic and accurate prostate gland segmentation on both DWI and T2WI images. This model was tested in 3 different external cohorts and showed satisfactory results on T2WI images. The segmentation performance on DWI images was inferior but still inspiring in the external testing group. This study might benefit the management of prostate diseases in the future.
Introduction
Accurate prostate whole gland segmentation on MR images plays an important role in the management of prostate cancer and even benign prostate hyperplasia, which is critical for MR-ultrasound fusion biopsy, cancer staging, and radiation treatment planning [1,2]. However, manual segmentation is a time-consuming process. Recently proposed U-Net-based deep learning convolutional neural network (CNN) architecture has been successfully applied to prostate segmentation [3]. Nevertheless, most of these studies were based on public datasets, and only a fraction of them tested their models in multi-center or multi-vendor cohorts [4,5]. Besides, a few models were designed for multiparametric MR images. Our study aimed to develop a 3D UNet model for automatic and accurate prostate whole gland segmentation which is reliable on both diffusion-weighted imaging (DWI) and T2-weighted imaging (T2WI) images.Methods
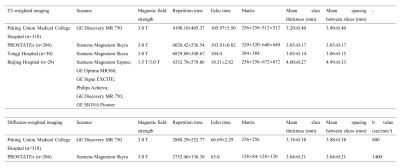
This study retrospectively enrolled 318 patients who underwent multiparametric prostate MRI and subsequent biopsy between November 2014 to December 2018 at our institution. These patients were divided into the training group (223 patients) and the independent internal testing group (95 patients) according to their examination time. To verify the performance of this model, 3 cohorts from different centers with different vendors were collected. Specifically, ninety patients from the test split of the PROSTATEx Challenge dataset were included as the external testing group 1 (E1). Thirty patients from Tongji Hospital and 29 patients from Beijing Hospital were included as external testing group 2 (E2) and external testing group 3 (E3), respectively. The detailed MRI acquisition parameters of each group were presented in Table 1. The patients’ axial T2WI and DWI (b value=800 sec/mm² for our institution, and 1400 sec/mm² for the PROSTATEx dataset) were collected and manually segmented by one radiologist to serve as the ground truth, another senior radiologist reviewed the images and modified the contour if necessary. For groups E2 and E3, only T2WI images were available for testing because of the retrospective collecting manner. A 3D UNet CNN-based segmentation model was trained on the training group by self-configuring nnUnet, in which the optimal super-parameters were obtained automatically. For each fold of the nnUnet, the model was trained for 500 epochs with the initial learning rate of 0.01. Considering the large bias between the DWI images of group E1 and the training cohort caused by the b value, the model was finetuned on the external finetune group (using 196 patients from the train split of the PROSTATEx dataset) for 50 epochs with a lower learning rate of 0.001. The Dice similarity coefficient (DSC) and 95% Hausdorff distance (95HD) were used for model performance evaluation.Results
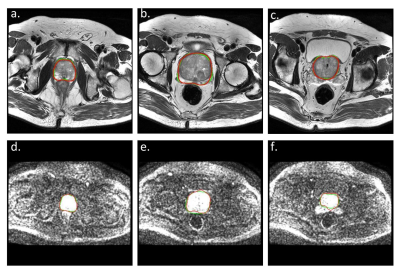
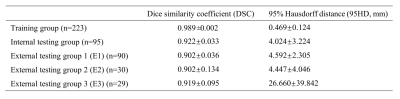
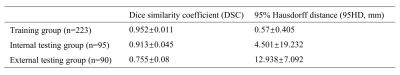
On T2WI, the average DSCs of 3D UNet were 0.989±0.002 in the training group and 0.922±0.033 in the internal testing group. In the three external testing groups, the average DSCs were also satisfactory (0.902 for E1 and E2, and 0.919 for E3). And the average 95HD in the testing groups was also satisfying (4.024, 4.592, and 4.447 mm for the internal testing group, E1, and E2, respectively), except for E3 (26.660 mm) (Table 2). And on DWI, the average DSCs were lower but still satisfying, reached 0.913±0.045 in the internal testing group, and were 0.755±0.08 in group E1. The averaged 95HD was 4.501 mm in the internal testing group and 12.938 mm in E1 (Table 3). Figure 1 showed an example of the 3D UNet model’s segmentation results compared with the radiologist’s manual segmentation.Discussion
In this study, we developed a robust deep learning CNN model to automatically segment the prostate whole gland on both T2WI and DWI images, and tested the model’s reliability in external testing groups. Segmentation results based on T2WI images were more stable and reliable, regardless of the differences in MR scanners and MR acquisition parameters. Comparatively, the segmentation performance on DWI images was more likely to be affected by acquisition parameters. Therefore, some improvement methods might be necessary for better segmentation of functional MRI images.Conclusion
The 3D UNet CNN model can automatically segment the prostate organ on both T2WI and DWI images with good performance, which might be helpful in the future segmentation of prostate cancer and benefitting the management of prostate cancer and benign prostate hyperplasia.Acknowledgements
No acknowledgement found.References
[1] Ghose S, Oliver A, Marti R, et al. A survey of prostate segmentation methodologies in ultrasound, magnetic resonance and computed tomography images. Comput Methods Programs Biomed, 2012, 108(1): 262-287.
[2] Almeida G, Tavares J. Deep Learning in Radiation Oncology Treatment Planning for Prostate Cancer: A Systematic Review. J Med Syst, 2020, 44(10): 179.
[3] Ushinsky A, Bardis M, Glavis-Bloom J, et al. A 3D-2D Hybrid U-Net Convolutional Neural Network Approach to Prostate Organ Segmentation of Multiparametric MRI. AJR Am J Roentgenol, 2021, 216(1): 111-116.
[4] Zavala-Romero O, Breto AL, Xu IR, et al. Segmentation of prostate and prostate zones using deep learning : A multi-MRI vendor analysis. Strahlenther Onkol, 2020, 196(10): 932-942.
[5] Soerensen SJC, Fan RE, Seetharaman A, et al. Deep Learning Improves Speed and Accuracy of Prostate Gland Segmentations on Magnetic Resonance Imaging for Targeted Biopsy. J Urol, 2021, 206(3): 604-612.
Figures