3659
Combine nnU-Net and radiomics for automated classification of breast lesion using mp-MRI
Jing Zhang1, Chenao Zhan2, Xu Yan3, Yang Song3, Yihao Guo4, Tao Ai2, and Guang Yang1
1East China Normal University, Shanghai key lab of magnetic resonance, shanghai, China, 2Tongji Medical College, Huazhong University of Science and Technology, Department of Radiology, Tongji Hospital, Wuhan, Hubei Province, China, 3Siemens Healthcare, MR Scientific Marketing, shanghai, China, 4Siemens Healthcare, MR Collaboration, Guangzhou, China
1East China Normal University, Shanghai key lab of magnetic resonance, shanghai, China, 2Tongji Medical College, Huazhong University of Science and Technology, Department of Radiology, Tongji Hospital, Wuhan, Hubei Province, China, 3Siemens Healthcare, MR Scientific Marketing, shanghai, China, 4Siemens Healthcare, MR Collaboration, Guangzhou, China
Synopsis
Multi-parametric MRI (mp-MRI) radiomics can distinguish breast mass effectively, but requires breast lesion segmentation first, which is subjective and laborious for radiologists. To overcome this problem, we combined nnUnet and radiomics analysis as an automatic model for breast lesion classification. In the test cohort, the breast lesion segmentation model achieved mean dice of 0.835, and the classification model achieved an AUC of 0.891. We found that the nnU-Net can delineatey lesions accurately based on dynamic contrast-enhanced (DCE, TWIST-VIBEs)), and mp-MRI radiomics features extracted from the auto-segmented lesions can be used to classfy breast lesions accurately.
INTRODUCTION
Breast cancer is the most commonly diagnosed cancer in females, accounting for 17.1% of new cancer cases in 20191, 2. Multi-parametric MRI (mp-MRI) is commonly used for breast cancer detection, and previous studies showed that radiomics analysis achieved the highest sensitivity in identifying malignant breast cancers by using a standard protocol and kinetic maps3–5. However, most radiomics feature extraction relies on lesion segmentation, which is laborious and subjective. The nnU-Net has been recently proposed to automatically adapt preprocessing strategies and network architectures for a given medical image dataset, and have been successfully used in different datasets6. In this work, we combined a segmentation model based on nnU-Net with a radiomics model for automated classification of breast lesion, it would help radiologist to diagnosis accurately.METHODS
A total of 192 patients with 222 pathologically confirmed lesions (malignant 178/benign 44) were enrolled in this retrospective study. The dataset was randomly split into a training cohort (156) and a test cohort (66). T2W, diffusion-weighted imaging (DWI), apparent diffusion coefficient (ADC) map, dynamic contrast material-enhanced (DCE), and 6 kinetic maps were acquired for all cases. One radiologist with 5-years’ experience outlined the lesion on T1Wpost90s, onto which all other images were aligned. Breast regions were also outlined for 25 cases to build a breast segmentation model.Firstly, we trained a breast segmentation model with nnU-Net, to segment the breast region from T1Wpost 90s. Then T1Wpost 90s images were multiplied with a segmented mask before being used as input to a lesion segmentation model based on nnU-Net. Finally, radiomics features were extracted from the lesion mask created by the lesion segmentation model. We used an open-source software FeatureExplorer for radiomics feature extraction and model building to multi-parametric MRI images7. K-means clustering was used for eliminating repetitive features, and recursive feature elimination (RFE) was used for feature selection by 10-fold cross-validation in the training cohort. Both supported vector machine (SVM) and logistic regression (LR) were tried as classifiers. The model that yielded the best performance on the cross-validation was chosen as the candidate model. To evaluate the value of shape features to diagnosis, we built a model with only shape features. To compare diagnosis values of different sequences and quantitative maps, we built radiomics models for each of the sequences and quantitative kinetic maps. Then a combined ModelDCE was built with features selected in DCE-derived models (T1Wpre, T1Wpost90s, T1Wpost5min, and 6 kinetic maps), and a combined ModelnonDCE was built with features from ADC map, DWI and T2WI. Finally, all selected features were used to train a combined model, namely Modelall. The Dice score is used to evaluate the accuracy of segmentation. The performance of the classification model was evaluated with receiver operating characteristic (ROC) curve analysis and confusion matrix. The results of ModelDCE, ModelnonDCE, and Modelall were also compared with Breast Imaging Reporting and Data System (BIRADS) reported by radiologists (malignant for BIRADS >3 or BIRADS >4).
RESULTS
The breast and lesion segmentation models achieved mean dice coefficients of 0.976 and 0.835, respectively. Three random cases are shown in Figure 2 to illustrate the results of breast segmentation and lesion segmentation. 107 radiomics features were extracted from each of T1W, kinetic parameter maps, ADC map, T2W, and DWI images. The results of all models are listed in Table 1. Among all single-modality models, T1Wpost90s achieved the highest AUC of 0.820 in the test cohort, and most features retained in the model are texture features. The shape model achieved an AUC value of 0.775.The combined ModelDCE and ModelnonDCE achieved AUC values of 0.865 and 0.861 in the test cohort, respectively, better than the assessment from the BIRADS (0.791). Furthermore, Modelall, which was built from all features retained in single-modality models, achieved the highest test AUC value of 0.891. A total of 15 features were selected in Modelall, including 11 texture features and 4 first-order features from T1Wpre, ADC map, 2 kinetic maps, T2W and DWI. Texture features from kinetic maps contributed most in modelall.
DISSCUSION
It can be seen from both the mean Dice coefficient and the final classification performance that segmentation models achieved satisfying results for both breast and lesion segmentation. Compared with previous work, our breast lesion segmentation models achieved higher performance8, 9. This two-step segmentation scheme minimized the false positive in lesion segmentation and made the whole pipeline fully automated, producing more subjective results while reducing the workload for radiologists. The ModelT1Wpost90s achieved the highest AUC value in all single-modality models, indicating that T1Wpost90s has the highest value for diagnosis. The final combined model achieved a test AUC of 0.891, higher than previous studies10, 11, proved that useful information can be extracted from various MRI sequences and quantitative maps. Thus, mp-MRI can be used to diagnose breast cancer effectively. The weakness of our work is poor segmentation ability for small lesions, this disadvantage leads to lower classification ability based on radiomics.CONCLUSION
By combining segmentation models with radiomics classification model, we constructed a fully automated pipeline for breast lesion diagnosis on mp-MRI and achieved satisfying performance.Acknowledgements
No acknowledgement found.References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018. GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018:68:394–424.
- Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: a cancer journal for clinicians 2021.
- Mann RM, Cho N, Moy L. Breast MRI. State of the Art. Radiology 2019:292:520–536.
- Parekh VS, Jacobs MA. Integrated radiomic framework for breast cancer and tumor biology using advanced machine learning and multiparametric MRI. NPJ Breast Cancer 2017:3:43.
- Zhang Q, Peng Y, Liu W, et al. Radiomics Based on Multimodal MRI for the Differential Diagnosis of Benign and Malignant Breast Lesions. Journal of magnetic resonance imaging : JMRI 2020:52:596–607.
- Fabian Isensee, Paul F. Jaeger, Simon A. A. Kohl, et al. nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation.
- Song Y, Zhang J, Zhang YD, et al. FeAture Explorer (FAE). A tool for developing and comparing radiomics models. PLoS One 2020:15:e0237587.
- Zhang J, Saha A, Zhu Z, et al. Hierarchical Convolutional Neural Networks for Segmentation of Breast Tumors in MRI With Application to Radiogenomics. IEEE transactions on medical imaging 2019:38:435–447.
- Qiao M, Suo S, Cheng F, et al. Three-dimensional breast tumor segmentation on DCE-MRI with a multilabel attention-guided joint-phase-learning network. Computerized medical imaging and graphics : the official journal of the Computerized Medical Imaging Society 2021:90:101909.
- Qiao M, Li C, Suo S, et al. Breast DCE-MRI radiomics: a robust computer-aided system based on reproducible BI-RADS features across the influence of datasets bias and segmentation methods. International journal of computer assisted radiology and surgery 2020:15:921–930.
- Feng H, Cao J, Wang H, et al. A knowledge-driven feature learning and integration method for breast cancer diagnosis on multi-sequence MRI. Magnetic resonance imaging 2020:69:40–48.
Figures
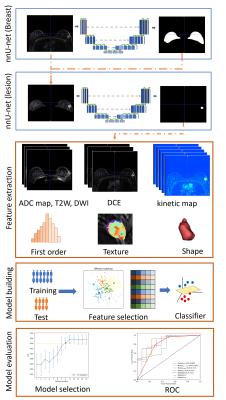
Figure1.Workflow
of this study. Segmentation models based on nnU-Net were trained for breast and
lesion segmentation. First-order and texture features were extracted from lesion
shapes and each of the sequences and quantitative maps to build radiomics
models. Finally, models were evaluated with ROC analysis on the test cohort.
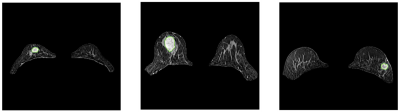
Figure2.nnU-net
segmentation result.
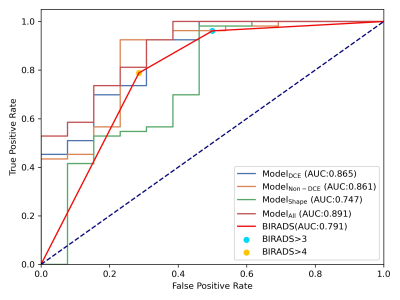
Figure3.ROC of the combined model in the test cohort.
Modelall has the highest AUC value. AUC value of Modelall,
ModelDCE, Modelnon-DCE are higher than Modelshape
and BIRADS.
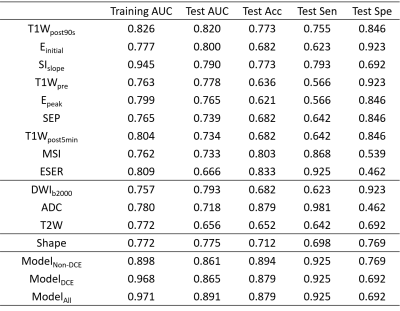
Table 1.The
clinical statistics of the different models on the training and test cohort.
DOI: https://doi.org/10.58530/2022/3659