3654
Comparison of BOLD detectability of ‘multi shot EPI’ and ‘single shot EPI with GRAPPA’ at 7T
Guoxiang Liu1,2, Adnan Shah1,2, Takashi Ueguchi1,2, and Seiji Ogawa1,3
1NICT, Osaka, Japan, 2Graduate School of Frontier Biosciences, Osaka University, Osaka, Japan, 3Tohoku Fukushi University, Sendai, Japan
1NICT, Osaka, Japan, 2Graduate School of Frontier Biosciences, Osaka University, Osaka, Japan, 3Tohoku Fukushi University, Sendai, Japan
Synopsis
We compare the detectability of multi shot EPI (msEPI) and single shot EPI with GRAPPA (ssEPI-GRAPPA) using D-value maps to show the benefit of BISEPI (Block-Interleaved Segmented EPI) [1] for the same total acquisition time. D-values were derived based on the relationship between target effect, the significance level of detection, the number of time points (volumes), and tSNR [2]. The proposed D-value maps can be a useful quality assurance measure for determining the tSNR gain and activity detection performance of msEPI in comparison to ssEPI-GRAPPA in high-resolution fMRI at 7T.
Introduction
We previously introduced a new multi shot EPI (msEPI) technique called block-interleaved segmented EPI (BISEPI) [1]. It realigns the k-space trajectories according to the timing information of the stimuli originating from a block design paradigm during acquisition and reconstruction to maintain the temporal resolution and temporal SNR (tSNR). In this work, we discuss BOLD detectability to show the benefit of BISEPI comparing to normal single shot EPI with GRAPPA (ssEPI-GRAPPA) for the same total acquisition time. One phantom data set acquired at 1.0-mm isotropic resolution and two human subject data sets acquired at 0.7-mm isotropic resolution showed that BISEPI is one choice to detect BOLD response at submillimeter high-resolution in fMRI studies.Materials & Methods
Based on the relationship between target effect, the significance level of detection, the number of time points (volumes), and tSNR [2], we defined D = TGRAPPA/Tms that indicates the detectability of ssEPI-GRAPPA compared with msEPI for the same acquisition time. TGRAPPA and Tms are total acquisition time needed to reach the same BOLD detectability using ssEPI-GRAPPA and msEPI separately. Hence, D > 1 indicates that ssEPI-GRAPPA requires a longer acquisition time to reach the same detectability of msEPI. Using the same TR value for single-shot imaging , we have $$D=\frac{T_{GRAPPA}}{T_{ms}}=\frac{N_{GRAPPA}}{(N_{ms})(R)}=(\frac{tSNR_{ms}}{tSNR_{GRAPPA}})^2(\frac{1}{R})$$ where NGRAPPA is the number of acquired volumes in ssEPI-GRAPPA with the GRAPPA acceleration factor R, and Nms is the number of reconstructed volumes in msEPI. Phantom and human subject’s data were acquired on a Siemens MAGNETOM 7T scanner with a 32-channel phased array head coil (Nova Medical, MA, USA). We calculated the D-values for a phantom scanned using ssEPI-GRAPPA for R values of 2, 3, and 4 and msEPI with 2, 3, and 4 segments at two slab locations and orientations. The major imaging parameters included 3D EPI sequence, 16 slices per slab, TR = 2000 ms, echo time of 32.3 ms, resolution of 1.0 mm isotropic, partial Fourier factor of 6/8, and flip angle of 21°. The GRAPPA acceleration factor R in ssEPI-GRAPPA matched the number of segments in msEPI for each calculation of D. The D-values are denoted as ssEPI-msEPI-x, where x is either GRAPPA acceleration factor R or the number of segments. Based on the computed D-values, slice maps of the phantom were generated for ssEPI-msEPI-2, ssEPI-msEPI-3, and ssEPI-msEPI-4, as shown in Figure 1. The D-values were calculated for two healthy subjects, whose scans were acquired using ssEPI-GRAPPA-3 and msEPI-3 (msEPI with 3 segments). The imaging parameters are: 24 slices per slab, TR = 1500 ms, echo time of 19.3 ms, FOV read of 45 mm, FOV phase of 90 mm, phase for partial Fourier factor of 6/8, flip angle of 25°, bandwidth of 1002 Hz/pixel, and oblique coronal slab orientation. Moreover, one subject was scanned using the same parameters under 18s/12s ON/OFF flipping checkerboard visual stimuli of total 10 minutes.Results and Discussion
Phantom time-series data are usually dominated by thermal image noise, and fluctuations is relatively small. The D-value difference observed in Figure 1 for ssEPI-msEPI-2, ssEPI-msEPI-3, and ssEPI-msEPI-4 predominantly arises from the geometric factor (g-factor) difference (columns in Figure 1), which strongly depends on the location of the imaging volume (rows in Figure 1). The large D-values obtained from the data of the two human subjects acquired using ssEPI-GRAPPA-3 and msEPI-3 at 0.7-mm isotropic resolution (Figure 2) are caused by the low tSNR of ss-EPI-GRAPPA-3 compared with msEPI-3. Hence, ssEPI-GRAPPA-3 may have a higher influence of fluctuations and g-factor, resulting in a lower sensitivity for activity detection compared with msEPI-3 for acquisition at these locations. The slice D-value maps and detected activity maps of the subjects are shown in Figure 2 and Figure 3, respectively. The proposed D-values and fMRI experimental results demonstrate the benefit of BISEPI, a kind of ‘multi shot EPI’ compared to ‘single shot EPI with GRAPPA’ for performing submillimeter high-resolution fMRI at 0.7-mm isotropic.Acknowledgements
No acknowledgement found.References
1. Liu G, Shah A, Ueguchi T. Block-Interleaved Segmented EPI for voxel-wise high-resolution fMRI studies at 7T. Proc. Intl. Soc. Mag. Reson. Med. 26; 2018; 5450.2. Murphy K, Bodurka J, Bandettini PA. How long to scan? The relationship between fMRI temporal signal to noise ratio and necessary scan duration. Neuroimage. 2007;34:565–574. doi: 10.1016/j.neuroimage.2006.09.032
Figures
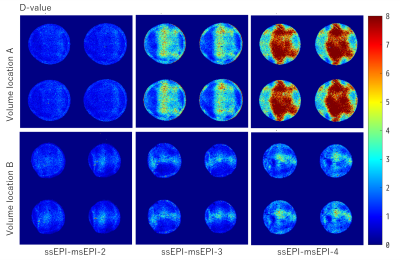
Figure 1: D-value maps for phantom scanned using ssEPI-GRAPPA and msEPI, where GRAPPA acceleration factor R in ssEPI-GRAPPA was matched with the number of segments in msEPI: ssEPI-msEPI-2, ssEPI-msEPI-3, and ssEPI-msEPI-4.
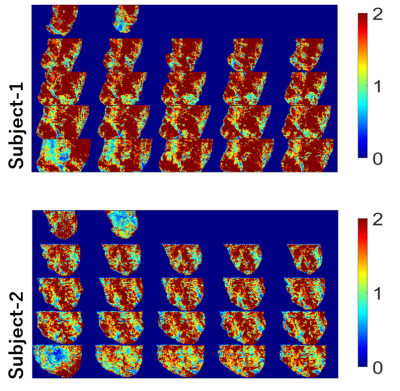
Figure 2: D-value maps for two subjects after motion correction based on images. The datasets were acquired using ssEPI-GRAPPA-3 and msEPI-3. msEPI-3 data were realigned using the proposed BISEPI [1].
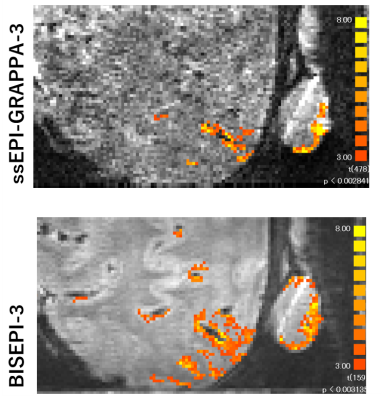
Figure 3: Detected fMRI activity during visual tasks of viewing flipping checkerboard using ssEPI-GRAPPA-3 (top) and BISEPI-3 (bottom). BISEPI-3 refers to BISEPI with 3 segments.
DOI: https://doi.org/10.58530/2022/3654