3624
A radiomics signature-based nomogram to predict TERT promoter mutation status and the prognosis of glioblastoma
Jun Lu1, Hailiang Li2, and Xiang Li1
1Department of Radiology, Affiliated Cancer Hospital of Zhengzhou University & Henan Cancer Hospital, Zhengzhou, China, 2Department of Radiology and Intervention, Affiliated Cancer Hospital of Zhengzhou University & Henan Cancer Hospital, Zhengzhou, China
1Department of Radiology, Affiliated Cancer Hospital of Zhengzhou University & Henan Cancer Hospital, Zhengzhou, China, 2Department of Radiology and Intervention, Affiliated Cancer Hospital of Zhengzhou University & Henan Cancer Hospital, Zhengzhou, China
Synopsis
This study aimed to establish and validate a radiomics signature-based nomogram with robust radiomics features from contrast enhanced MRI images. The radiomics features were selected using LASSO regression. A prediction model was constructed with multivariate logistic regression analysis. A nomogram combined radiomics signature and clinical factors were established, showing good performance for predicting the TERT mutation status. The clinical value of radiomics nomogram was further assessed by the prognosis analysis. In conclusion, the radiomics signature-based nomogram is a promising method for preoperatively predicting TERT promoter mutation status and has the potential to assess prognosis noninvasively in glioblastoma patients.
Introduction
Glioblastoma is the most common malignant primary central nervous system(CNS) tumor1. Molecular features were included in the 2016 World Health Organization(WHO) classification criteria for CNS tumors2,3. The telomerase reverse transcriptase(TERT) promoter has emerged as a strong prognostic and predictive biomarker in glioblastoma. Previous study found that an increase in matrix metalloproteinase expression would result in increased blood brain barrier(BBB) permeability4,5. The TERT mutation status can regulate the expression of metalloproteinases, which can affect the treatment results of drug6,7. In recent years, inhibition of TERT gene mutation is expected to become a new therapeutic target with the development of precision therapy8,9. TERT mutation status is expected to be routinely measured in the clinical evaluation. However, TERT mutation status is assessed invasively and subject to tumor heterogeneity through biopsy or surgical resection after comparatively long detection periods. Therefore, there is a strong need for noninvasive imaging biomarkers to predict TERT mutation status for making a tailored treatment plan precisely and prognosis assessment in glioblastoma from the initial stage of the tumor diagnosis.Radiomics, a recently emerging technique with high-throughput quantitative data, makes it possible for differential diagnosis and prognosis evaluation10. Several studies using the Visually Accessible Rembrandt Images(VASARI) and apparent diffusion coefficient(ADC) value failed to find the correlations between imaging features and TERT mutation status effectively11-13. Recently, promising results have revealed the use of radiomics signature to predict molecular characteristics such as IDH and MGMT, limited studies have focused on predicting TERT status14-16. Tian et al. and Arita et al. studied the radiomics signature for predicting TERT mutation of lower-grade gliomas and reported that the accuracy ranged from 0.56 to 0.74, demonstrating an unsatisfying result17,18. Since the TERT mutation status has been proven to be associated with treatment plan and prognosis assessment of glioblastoma8,19, prediction of the TERT mutation status using radiomics signature-based nomogram may have great clinical potential. Besides, few studies explored the correlations between radiomics signature and prognosis of glioblastoma. The purpose of this study was, therefore, to investigate the radiomics signature-based nomogram for noninvasively predicting the TERT promoter mutation status and prognosis in glioblastoma patients.
Methods
176 glioblastoma patients(123 in the training cohort and 53 in the validation cohort) were retrospectively enrolled. TERT promoter mutation(C228T and C250T) was determined with a Sanger sequencing. A total of 851 radiomics features were extracted from contrast enhanced MRI images using PyRadiomics software(Fig.1). The radiomics features were selected using the least absolute shrinkage and selection operator(LASSO) method(Fig.2) and a rad-score was calculated. A prediction model was constructed with multivariate logistic regression analysis. A nomogram combined radiomics signature and clinical factors were established(Fig.3). The performance of the model was evaluated by receiver operating characteristic(ROC) curve analysis. Kaplan-Meier curves were plotted based on the TERT promoter mutation status and the radiomics signature-based prediction model in stratifying the overall survival(OS) of patients. The log-rank test was utilized to determine differences in survival between the groups.Results
The rad-score was calculated with seven robust radiomics features(SurfaceArea, Skewness, Strength, SizeZoneNonUniformity, TotalEnergy, Imc1 and ZoneEntropy) showing good performance for predicting the TERT promoter mutation status. With multivariate logistic regression analysis, the radiomics signature was the independent predictor. The accuracy, sensitivity, specificity and AUC were 84.55%, 94.20%, 72.22%, 0.900(95% confidence interval [CI]: 0.832-0.946) in the training cohort and 83.02%, 82.14%, 84.00%, 0.873(95%CI: 0.753-0.948) in the validation cohort, respectively (Fig.4).With a median overall survival time of 18.5 months, both the TERT promoter mutation status and the radiomics signature stratified the glioblastoma patients into a high-risk group and a low-risk group (p < 0.001), and the differences within the high and low risk groups did not show statistical significance(p=0.140, p=0.381)(Fig.5).
Discussion
In this study, TERT promoter mutation status-related radiomics nomogram including the radiomics signature and clinical characteristics was established, showing good performance for predicting the TERT mutation status. The clinical value of the radiomics nomogram was further assessed by the prognosis analysis.The contrast enhanced MRI images can provide accurate information of angiogenesis representing tumor characteristics with clear boundaries. A major reason for the excellent prediction performance of radiomics nomogram may be the appropriate sequence, where contrast enhanced MRI-based radiomics features provide accurate information of angiogenesis thanks to the injection of gadolinium, thin slice thickness (1 mm) compared to previous studies with the slice thickness of 3.5-5.0 mm17,20.
In our study, the radiomics nomogram could stratify patients into two significantly different groups based on the prognosis, suggesting the feasibility of using the radiomics nomogram to predict prognosis in addition to predicting TERT mutation status. Moreover, the differences between the TERT mutation status-predicted and radiomics nomogram-predicted prognosis within each risk group showed no significance. The clinical use of a radiomics nomogram can be further supported since the radiomics nomogram can not only detect the TERT mutation status noninvasively but also predict the patients’ prognosis before treatment.
Conclusion
The radiomics signature-based nomogram with imaging characteristics from contrast enhanced MRI images is a promising method for preoperatively predicting the TERT promoter mutation status and has the potential to guide the treatment and predict the prognosis noninvasively in glioblastoma patients.Acknowledgements
NoneReferences
1.Louis D N, Perry A, Wesseling P, et al. The 2021 WHO Classification of Tumors of the Central Nervous System: a summary[J]. Neuro Oncol, 2021,23(8):1231-1251.2.Weller M, van den Bent M, Tonn J C, et al. European Association for Neuro-Oncology (EANO) guideline on the diagnosis and treatment of adult astrocytic and oligodendroglial gliomas[J]. Lancet Oncol, 2017,18(6):e315-e329.
3.Nabors L B, Portnow J, Ammirati M, et al. NCCN Guidelines Insights: Central Nervous System Cancers, Version 1.2017[J]. J Natl Compr Canc Netw, 2017,15(11):1331-1345.
4.Park Y J, Kim E K, Bae J Y, et al. Human telomerase reverse transcriptase (hTERT) promotes cancer invasion by modulating cathepsin D via early growth response (EGR)-1[J]. Cancer Lett, 2016,370(2):222-231.
5.Rempe R G, Hartz A, Bauer B. Matrix metalloproteinases in the brain and blood-brain barrier: Versatile breakers and makers[J]. J Cereb Blood Flow Metab, 2016,36(9):1481-1507.
6.Ichimura K. TERT promoter mutation as a diagnostic marker for diffuse gliomas[J]. Neuro Oncol, 2019,21(4):417-418.
7.Horn S, Figl A, Rachakonda P S, et al. TERT promoter mutations in familial and sporadic melanoma[J]. Science, 2013,339(6122):959-961.
8.Li X, Qian X, Wang B, et al. Programmable base editing of mutated TERT promoter inhibits brain tumour growth[J]. Nat Cell Biol, 2020,22(3):282-288.
9.Mancini A, Xavier-Magalhaes A, Woods W S, et al. Disruption of the beta1L Isoform of GABP Reverses Glioblastoma Replicative Immortality in a TERT Promoter Mutation-Dependent Manner[J]. Cancer Cell, 2018,34(3):513-528.
10.Lambin P, Leijenaar R, Deist T M, et al. Radiomics: the bridge between medical imaging and personalized medicine[J]. Nat Rev Clin Oncol, 2017,14(12):749-762.
11.Ivanidze J, Lum M, Pisapia D, et al. MRI Features Associated with TERT
Promoter Mutation Status in Glioblastoma[J]. J Neuroimaging, 2019,29(3):357-363.
12.Yamashita K, Hatae R, Hiwatashi A, et al. Predicting TERT promoter mutation
using MR images in patients with wild-type IDH1 glioblastoma[J]. Diagn Interv Imaging, 2019,100(7-8):411-419.
13.Park Y W, Ahn S S, Park C J, et al. Diffusion and perfusion MRI may predict EGFR amplification and the TERT promoter mutation status of IDH-wildtype lower-grade gliomas[J]. Eur Radiol, 2020,30(12):6475-6484.
14.Lu J, Li X, Li H. Perfusion parameters derived from MRI for preoperative prediction of IDH mutation and MGMT promoter methylation status in glioblastomas[J]. Magn Reson Imaging, 2021,83:189-195.
15.Jiang C, Kong Z, Liu S, et al. Fusion Radiomics Features from Conventional MRI Predict MGMT Promoter Methylation Status in Lower Grade Gliomas[J]. Eur J Radiol, 2019,121:108714.
16.Jiang C, Kong Z, Zhang Y, et al. Conventional magnetic resonance imaging-based radiomic signature predicts telomerase reverse transcriptase promoter mutation status in grade II and III gliomas[J]. Neuroradiology, 2020,62(7):803-813.
17.Tian H, Wu H, Wu G, et al. Noninvasive Prediction of TERT Promoter Mutations in High-Grade Glioma by Radiomics Analysis Based on Multiparameter MRI[J]. Biomed Res Int, 2020,2020:3872314.
18.Arita H, Kinoshita M, Kawaguchi A, et al. Lesion location implemented magnetic resonance imaging radiomics for predicting IDH and TERT promoter mutations in grade II/III gliomas[J]. Sci Rep, 2018,8(1):11773.
19.Ohba S, Kuwahara K, Yamada S, et al. Correlation between IDH, ATRX, and TERT promoter mutations in glioma[J]. Brain Tumor Pathol, 2020,37(2):33-40.
20.Fang S, Fan Z, Sun Z, et al. Radiomics Features Predict Telomerase Reverse Transcriptase Promoter Mutations in World Health Organization Grade II Gliomas via a Machine-Learning Approach[J]. Front Oncol, 2020,10:606741.
Figures
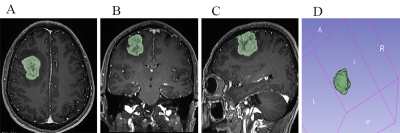
Fig. 1 (A-D) Male, 48 years, glioblastoma, TERT mutation. The green areas represent the regions of interest segmented on transverse(A), coronal(B), sagittal(C) section. Example of volume of interest (VOI) segmentation on T1-weighted magnetization prepared rapid acquisition gradient echo image(D).
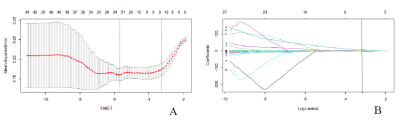
Fig. 2 Feature selection using LASSO regression. (A) LASSO coefficient profiles of the 40 selected features via 10-fold cross-validation. Two vertical lines were plotted with the minimum criteria and the 1 standard error of the minimum criteria. (B) A vertical line was plotted at the optimal λ value after calculating the mean squared error and goodness of fit, which resulted in 7 features with nonzero coefficients.
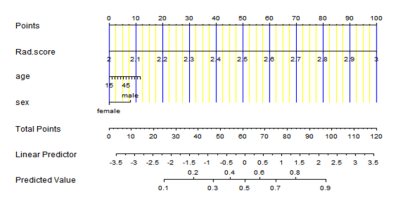
Fig. 3 The radiomics nomogram based the radiomics signature and clinical factors. The predicted values for predicting the TETRT promoter mutation were showed.
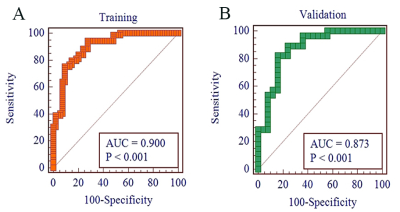
Fig. 4 Graph shows ROC curves of radiomics signature for predicting the TERT mutation status in the (A)training and (B)validation cohort.
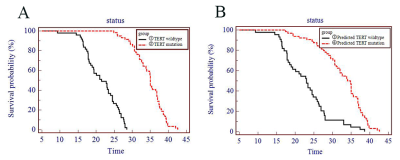
Fig. 5 Kaplan-Meier curves revealed the prognosis-based groups stratified by the TERT promoter mutation status(A) and the radiomics signature(B). (①vs②:p<0.001;③vs④:p<0.001; ①vs③:p=0.381; ②vs④:p=0.140)
DOI: https://doi.org/10.58530/2022/3624