3559
Towards fast single-slice QSM: Challenges and possible solutions1Department of Biomedical Engineering, University of Alberta, Edmonton, AB, Canada
Synopsis
Susceptibility maps reconstructed from small-axial slabs suffer underestimation due to background-field removal imperfections near slab boundaries and the increased difficulty of solving a 3D-inversion problem with reduced-support in the direction of B0. Reliable QSM reconstruction from small slabs would enable higher resolution imaging within a reasonable time-frame and further accelerate clinical adoption. This work proposed using additional rapid low-resolution data, that serves as prior information, to improve background-field estimation and regularize the inversion-to-susceptibility process. Consistent high-resolution QSM at 3T was obtained from slabs of width as small as 6.8mm, aided by a lower-resolution dataset of 16 times coarser voxels.
INTRODUCTION
Quantitative Susceptibility mapping (QSM) has evolved in the last decade to visualize and measure metallic deposits in human tissues, and has been used in many neurological studies. QSM is typically reconstructed from a whole brain 3D phase volume acquired via a multi-echo gradient-echo (MEGE) sequence. The necessity of collecting large 3D spatial coverage arises from the underlying 3D physical modelling that requires field information in the vicinity of the ROI. Previous studies have reported elevated underestimation in QSM when the Volume of Interest (VOI) is reduced, more noticeably in iron-rich regions, and suggested a minimum coverage of 2-6 times the ROI size for reliable measurement 1,2. Although fast large volume QSM is achievable using advanced parallel imaging techniques 3 and/or fast pulse sequences 4, producing QSM from smaller VOIs would further accelerate the acquisition, allow higher resolution imaging, and accelerate clinical adoption. Here, we explore the challenges incurred in reduced-VOI QSM applications and propose improving the reconstruction with the aid of additional rapid low-resolution data.METHODS
Imaging Protocol:Brain images from healthy subjects were collected at 3T after giving written informed consent and under the approval of the local ethics committee. The imaging protocol included: a) 0.87mm isotropic MPRAGE for segmentation, b) high-resolution (0.47x0.47x0.85 mm3) MEGE with 74.8mm through-plane coverage (88 slices acquired in 13 minutes), and c) low-resolution (0.94x0.94x3.4 mm3) MEGE with 149.6mm full head coverage (44 slices acquired in 1.5 minutes). Other parameters included: axial slices, 13° flip angle, 2 GRAPPA acceleration, 6 echoes, TE1: 4.62ms/3.82ms, ∆TE: 7.11ms/5.49ms and TR: 47ms/37ms for high/low resolution data. Additional high resolution MEGE volumes with smaller coverage were produced from (b) by limiting the number of slices to: 42, 20, 8, 4 and 2.
QSM Reconstruction:
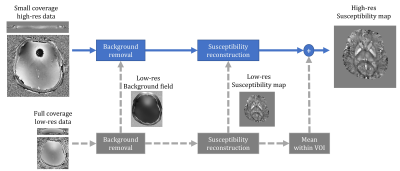
The proposed approach employed the additional low-res data to improve the reconstruction in three steps, as shown in Fig.1 (gray blocks and arrows). First, local field was estimated by subtracting the low-res background field (after interpolation). Then, low-res local field and/or susceptibility were/was used as a prior knowledge to regularize the inversion process. Here, the latter was used via L2-norm. Finally, the mean value of the low-res QSM within VOI can be used to correct the DC-term in the final high-res QSM.
Prior to any processing for the high-res data, the outer 4 slices were excluded due to poor SNR at the edge of the imaged slab. Common processing steps between the proposed and the standard approach (Fig.1 blue blocks and arrows) included: brain extraction using BET tool from FSL package 5, phase unwrapping and total field estimation using ROMEO 6, background field removal using V-SHARP with maximum 12mm kernel radius 7, and susceptibility inversion using MEDI 8.
Segmentation and Measurement:
Four deep grey matter structures (caudate, putamen, thalamus, and globus pallidus) were segmented from the T1w images using VolBrain 9, and their masks were mapped into MEGE space after registration using ANTs tool 10. Then, ROI measurements were pooled from both hemispheres and compared in susceptibility maps obtained from different slab widths.
RESULTS
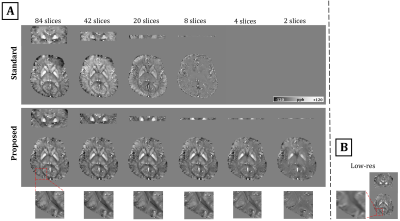
Fig.2A demonstrates the outputs of the two processing approaches. With the standard approach, susceptibility underestimation increased as brain coverage in the direction of main field decreased, and more than 4 slices were needed for successful reconstruction. In contrast, the proposed approach produced more consistent susceptibility values (Fig.2A & Fig.3) even with coverage as small as 6.8mm (8 slices). However, minimum coverage could be limited by field strength and SNR. Fig.2B illustrates the low-resolution QSM with x16 larger voxels used in the proposed approach.DISCUSSION
Imaging small slabs instead of full brain further reduces scanning time. Alternatively, saved time can be exploited to explore higher resolution (up to hardware/SNR limits) or improve SNR. However, accurate reconstruction of QSM from restricted spatial extension is challenging due to several processing issues. First, background-field removal methods are known for being increasingly less accurate toward VOI edges 11, and as the VOI shrinks the error in the resultant local field elevates substantially and propagates to the final susceptibility map. Second, the field-into-susceptibility inversion is an ill-conditioned 3D problem, and a global solution for the minimization formulation becomes more difficult as the support region for the solution decreases. Finally, solving this inversion problem yields a zero-mean solution within VOI, and thus the obtained QSM would be normalized by varying mean value depending on the VOI contents and coverage.We proposed using additional rapid low-res data to overcome the above-mentioned issues and stabilize the reconstruction solution. More consistent QSM was obtained with spatial coverage as small as 6.8 mm with aid from a dataset with x16 coarser resolution. Ultimately, the application of this approach would utilize the phase of any low-resolution GRE-based data already available in the imaging protocol, such as a field map, and thus minimize the burden of acquiring additional data. In addition, obtaining the local field by subtracting the low-res background field may enable QSM from even a single-slice.
Acknowledgements
This work was supported by the Natural Sciences and Engineering Research Council of Canada.References
1. Elkady A M., Sun H, Wilman A H. Importance of extended spatial coverage for quantitative susceptibility mapping of iron-rich deep gray matter. Magnetic resonance imaging. 2016; 34(4):574-578.
2. Karsa A, Punwani S, Shmueli K. The effect of low resolution and coverage on the accuracy of susceptibility mapping. Magnetic resonance in medicine. 2019; 81(3):1833-1848.
3. Bilgic B, Cauley S F, Fan A P, et al. Wave-CAIPI enables highly accelerated 3D MRI. 40th Annual Northeast Bioengineering Conference (NEBEC), 2014:1-2.
4. Sun H, Wilman A H. Quantitative susceptibility mapping using single‐shot echo‐planar imaging. Magnetic resonance in medicine. 2015; 73(5):1932-1938.
5. Smith S M. Fast robust automated brain extraction. Hum. Brain Mapp. 2002;17:143–155.
6. Dymerska B, et al. Phase unwrapping with a rapid opensource minimum spanning tree algorithm (ROMEO). Magnetic Resonance in Medicine. 2021; 85(4):2294-2308.
7. Li W, Wu B, Liu C. Quantitative susceptibility mapping of human brain reflects spatial variation in tissue composition. Neuroimage. 2011; 55:1645–1656.
8. Liu T, Wisnieff C, Lou M, et al. Nonlinear formulation of the magnetic field to source relationship for robust quantitative susceptibility mapping. Magn. Reson. Med. 2013; 69:467–476.
9. Manjón J V, Coupé P. Volbrain: An online MRI brain volumetry system. Front. Neuroinform. 2016;10.
10. Avants B B, Tustison N J, Song G, et al. A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage. 2011; 54:2033–2044.
11. Schweser F., Robinson S D, de Rochefort L, et al. An illustrated comparison of processing methods for phase MRI and QSM: removal of background field contributions from sources outside the region of interest. NMR Biomed. 2017; 30(4):e3604.
Figures