3523
Preserving Privacy While Maintaining Consistent Postprocessing Results: Fast and Effective Anonymous Refacing using a 3D cGAN1Advanced Clinical Imaging Technology, Siemens Healthcare AG, Lausanne, Switzerland, 2Signal Processing Laboratory (LTS 5), Ecole Polytechnique Fédérale de Lausanne (EPFL), Lausanne, Switzerland, 3Department of Radiology, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland
Synopsis
We propose a novel refacing technique employing a 3D conditional generative adversarial network to allow protecting subject privacy while maintaining consistent post-processing results. We evaluate the method compared to current refacing techniques using brain morphometry as an example. Results show that the proposed method compromises brain morphometry results to a lesser extent than existing methods while showing lower similarity of the final image to the original one, hence suggesting an improved privacy protection. We conclude that the proposed method represents a fast and viable alternative for image data de-identification compared to currently existing methods.
Introduction
Identification of individuals based on 3D renderings obtained from routine structural MRI scans of the head is a growing privacy concern. A recent study reported correct matching of 97% of participants’ MRI scans to photos1. For this reason, several algorithms have been developed to de-identify imaging data using blurring, defacing or refacing1-5. While completely removing facial structures provides the best re-identification protection, it can significantly affect subsequent analysis steps, such as brain morphometry4. On the other hand, refacing methods1,3,4 may provide consistent image processing results4,5, but with a higher risk of re-identification. Another common problem among the proposed methods is a long runtime, which can be a hurdle when processing large datasets.To address these issues, we propose a 3D conditional generative adversarial network (cGAN) for fast and effective anonymous refacing of defaced data. We investigated the impact of the approach on image processing algorithms compared to common refacing techniques using brain morphometry as an example application and analyzed different image similarity metrics as surrogate measure for subject identifiability.
Methods
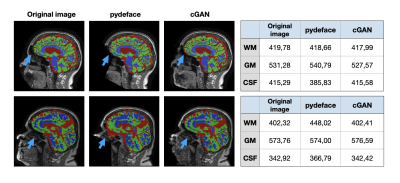
408 3D T1-weighted MRI scans of 102 Alzheimer’s Disease and mild cognitive impaired patients and 102 healthy controls were extracted from the TADPOLE dataset6 from ADNI7. All scans were reoriented to ASL orientation. Defaced images were generated using pydeface2 and qualitatively controlled for successful defacing.The proposed refacing technique consists of a 3D cGAN proposed by Cirillo et al.8 with an L1.5 generator loss term9. The cGAN was trained on 128x128x128 sub-volumes extracted from each image and combined in one batch. Fifty-two patient and fifty-two control pairs of original and defaced images were used for training and validation. Training was performed on an NVIDIA V100 GPU. The remaining 200 images were left for comparison of different refacing techniques and will be further referred to as the “test set”.
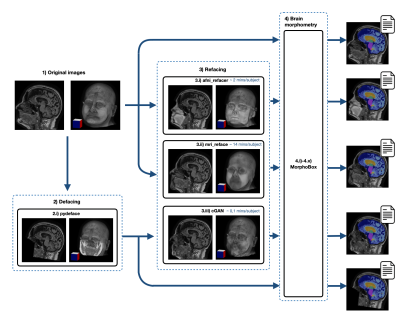
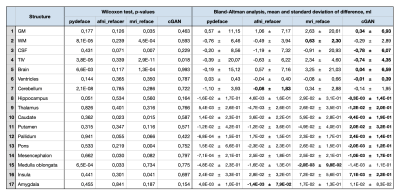
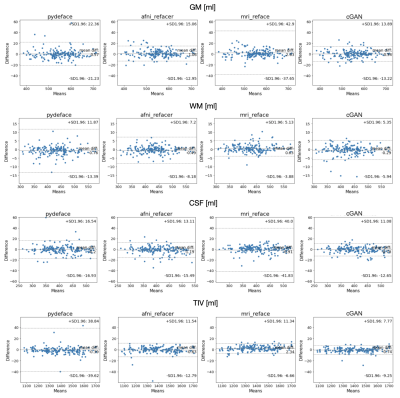
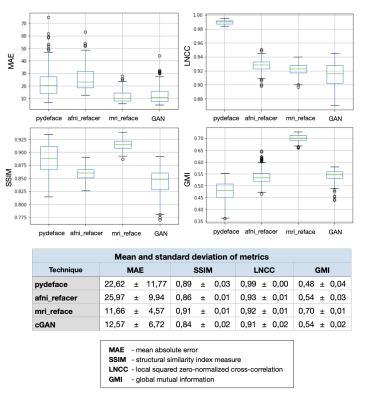
For evaluation, three different refacing tools were applied to the test set: i) afni_refacer3, ii) mri_reface1, iii) the trained cGAN. For cGAN’s predictions, the voxels within the head that had not been removed by defacing were replaced by their original values to avoid unwanted changes in the brain. Subsequently, volumes of brain structures were computed using the prototype software MorphoBox10 for i) original images, ii)-iv) refacing techniques’ outputs and v) defaced images. The processing workflow is illustrated in Figure 1. Eighteen subjects were excluded from further analysis, either because pydeface partially removed brain tissue or MorphoBox failed on the original images. Significant differences between absolute volumetric estimates of de-/refaced images and original images were assessed using paired Wilcoxon tests; an additional Bland-Altman analysis was performed for the main brain structures. Approximating the re-identification risk, several image similarity metrics were evaluated for pairs of de-/refaced and original images, that include: structural similarity index (SSIM), mean absolute error (MAE), local cross-correlation (LNCC) and global mutual information (GMI).
Results
Comparing the average runtime of the different methods on eight scans, our cGAN approach took 5 sec/scan on a GPU, outperforming both mri_reface and afni_refacer taking 14 and 2 min/scan, respectively. Statistical analysis of the volumetric results of 17 brain structures for different techniques is presented in Table 1. Bland-Altman plots for the main brain structures are presented in Figure 2. The volumes of 5 of 17 brain structures were significantly different ($$$p$$$-$$$value\leq0.05$$$) for pydeface output images compared to the original images; for afni_refacer 3/17 structures, for mri_reface 8/17 structures, and for cGAN 1/17 structures showed significant differences in volume. The cGAN refacing approach was able to mitigate statistically significant differences in volumetric results induced by defacing in all structures except the total intracranial volume (TIV) (Table 1, Figure 4). Means and standard deviations of volumetric differences with original images were the lowest for the cGAN output in most of the structures comparing to other techniques (Table 1, Figure 2). mri_reface showed comparable or worse performance than pydeface in 12/17 structures with Bland-Altman analysis (Table 1). The distributions of image similarity metrics for pairs of original and de-/refaced images are presented in Figure 3. The techniques that showed the lowest mean values for the respective metrics were: for SSIM and LNCC – cGAN, for MAE – mri_reface, for GMI – pydeface. The techniques that showed the highest metrics’ mean values: for SSIM and GMI – mri_reface, for MAE – afni_refacer, for LNCC – pydeface.Discussion and conclusions
We proposed a new refacing technique based on a 3D cGAN to protect subject privacy while ensuring consistent post-processing results. The performance of the method was compared to current refacing techniques using brain volumetric measurements to quantify the impact on image analysis algorithms. In contrast to other refacing techniques, the proposed approach did not result in significant volumetric differences compared to the original images except for the TIV.Evaluating image similarity metrics between de-/refaced and original images, we approximated the re-identification risk of regenerated faces. While the cGAN showed the lowest risk of re-identification, the expressive power of these metrics as surrogates for subject identifiability is certainly limited. Future work should include evaluation of face recognition tools to better evaluate this risk.
In conclusion, 3D cGANs provide a viable way for fast and robust subject image de-identification to preserve privacy while maximizing data reusability.
Acknowledgements
Data collection and sharing for this project was funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.References
1. Schwarz CG, Kremers WK, Wiste HJ, et al. Changing the face of neuroimaging research: Comparing a new MRI de-facing technique with popular alternatives. NeuroImage 2021, 231. doi: 10.1016/j.neuroimage.2021.117845.
2. Poldrack RA. Pydeface. From https://github.com/poldracklab/pydeface.
3. AFNI. afni_refacer. From https://github.com/PennLINC/afni_refacer.
4. Huelnhagen T, et al. Don’t Lose Your Face - Refacing for Improved Morphometry. ISMRM 2020.
5. Theyers AE, Zamyadi M, O'Reilly M, et al. Multisite Comparison of MRI Defacing Software Across Multiple Cohorts. Frontiers in Psychiatry 2021, 12:189. doi: 10.3389/fpsyt.2021.617997.
6. TADPOLE. https://tadpole.grand-challenge.org constructed by the EuroPOND consortium http://europond.eu funded by the European Union’s Horizon 2020 research and innovation programme under grant agreement No 666992.
7. Alzheimer’s Disease Neuroimaging Initiative (ADNI).
8. Cirillo MD, Abramian D, Eklund A. Vox2Vox: 3D-GAN for Brain Tumour Segmentation. BrainLes 2020. Lecture Notes in Computer Science, 12658. Springer, Cham. doi: 10.1007/978-3-030-72084-1_25.
9. Liu Y, Lei Y, Wang T, et al. CBCT-based synthetic CT generation using deep-attention cycleGAN for pancreatic adaptive radiotherapy. Med. Phys. 2020, 47(6):2472–2483. doi: 10.1002/mp.1412.
10. Schmitter D, Roche A, Maréchal B, et al. An evaluation of volume-based morphometry for prediction of mild cognitive impairment and Alzheimer’s disease. NeuroImage Clin. 2015;7:7–17 doi: 10.1016/j.nicl.2014.11.001.
Figures