3520
A Ontology-guided Attribute Partitioning Ensemble Learning Model for Early Prediction of Cognitive Deficits using sMRI in Very Preterm Infants1Imaging Research Center, Cincinnati Children's Hospital Medical Center, Cincinnati, OH, United States, 2Electrical Engineering and Computer Science, University of Cincinnati, Cincinnati, OH, United States, 3The Perinatal Institute, Cincinnati Children's Hospital Medical Center, Cincinnati, OH, United States, 4Pediatrics, University of Cincinnati, Cincinnati, OH, United States, 5Radiology, Cincinnati Children's Hospital Medical Center, Cincinnati, OH, United States
Synopsis
Between 35-40% of very preterm infants (≤32 weeks’ gestational age) develop cognitive deficits, thereby increasing their risk for poor educational, health, and social outcomes. Timely and accurate identification of infants at risk soon after birth is desirable for early intervention allocation. We proposed a novel Ontology-guided Attribute Partitioning Ensemble Learning (OAP-EL) model using quantitative structural MRI data obtained soon after birth to predict cognitive deficits at 2 years corrected age in very preterm infants.
INTRODUCTION
The prevalence of neurodevelopmental impairments remains high for very preterm infants (gestational age ≤32 weeks). Between 35-40% of very preterm infants (≤32 weeks’ gestational age) develop cognitive deficits. Yet, an accurate clinical diagnosis of cognitive deficits is currently unavailable for very preterm infants until 3-5 years of age in early childhood. The absence of prompt treatment leads to missing the optimal neuroplasticity period of brain development when interventions can exert the greatest impact on prevention.Multiple structural brain magnetic resonance imaging (sMRI) studies have shown that brain anatomical deformities might be associated with cognitive deficits in preterm infants (2-5). Ensemble learning (EL) has been frequently applied to improve the performance of machine learning models in a variety of medical applications (6-10). Recently, domain knowledge expressed in ontologies is increasingly being utilized in biomedical research studies, (11-13). In this study, we proposed to apply domain knowledge ontologies to guide sMRI feature partitioning and developed a novel Ontology-guided Attribute Partitioning Ensemble Learning (OAP-EL) model using quantitative sMRI features obtained at birth to predict cognitive deficits at 2 years corrected age in very preterm infants.
METHODS
In this IRB-approved work, we included 261 very preterm infants from five Cincinnati Ohio neonatal intensive care units (NICUs) (referred to as the CINEPS cohort); and 108 very preterm infants from four Columbus Ohio NICUs (referred as COEPS cohort). All subjects of CINEPS cohort were imaged at 39-44 weeks postmenstrual age (PMA) during unsedated sleep on a 3T Philips Ingenia scanner with a 32-channel receiver head coil. Acquisition parameters for axial T2-weighted fast spin-echo imaging parameters were repetition time (TR) = 8300 ms, echo time (TE) = 166 ms, flip angle (FA) = 90°, resolution 1.0 × 1.0 × 1.0 mm3: time 3:53 min. All COEPS subjects were scanned at 38-43 weeks PMA on a 3T MRI scanner (Skyra; Siemens Healthcare) using a 32-channel head coil. Acquisition parameters for axial T2-weighted fast spin-echo sequence were TR = 9500 ms, TE = 147 ms, FA = 150°, resolution 0.93 × 0.93 × 1.0 mm3; time 4:09 min.Each subject was assessed at 2 years corrected age using the well-established Bayley Scales of Infant and Toddler Development III (Bayley III) test. The Bayley-III cognitive scores ranged from 55 to 145 for the CINEPS cohort and 55 to 135 for the COEPS cohort. Using a threshold of 85, we dichotomized very preterm infants into two groups: low-risk infants (>85) vs. high-risk infants (≤85) to develop moderate/severe cognitive deficits. Detailed demographics of the subjects are listed in Table 1.
We preprocessed T2-weighted MRI data of each subject and extracted brain geometric features using the developing Human Connectome Project (dHCP) structural pipeline (14). The dHCP structural pipeline performed: bias field correction (17), brain extraction (18,19), brain surface reconstruction (21), and whole brain image segmentation into 87 ROIs guided by an age-matched neonatal volumetric atlas (18, 20). For individual subjects, the dHCP pipeline calculated a total of 510 brain geometric features, which can be categorized into six different types, including volume, thickness, sulcal depth, curvature, gyrification index, and surface area.
Figure 1 demonstrates the schematic diagram of the OAP-EL model. We first extract brain geometric features from T2w MRIs for individual very preterm infants using the dHCP pipeline (14) (Figure 1A). Next, based on 2 prior defined ontologies (14, 15), we construct an ontology graph, in which brain geometric features were considered as vertices and ontology-derived relationships were edges; and we conduct ontology graph clustering (Figure 1B) to partition brain geometric features into k non-overlapping feature subsets (Figure 1C). With each independent feature subset, we trained k base-models (i.e., eXtreme Gradient Boosting (XGBoost) classifiers (16) and then utilized a neural network model as a meta-model to integrate k individual base-models for risk stratification (Figure 1D). We internally validate our model with a leave-one-out cross-validation using CINEPS cohort, and test the internally validated model on the unseen independent COEPS cohort.
RESULTS
In internal validation, the OAP-EL model achieved a mean accuracy of 71.3%, a sensitivity of 70.6%, specificity of 72.6%, and an AUC of 0.74, outperforming both traditional machine learning (Figure 2) and peer ensemble learning models (Table 2). The OAP-EL model achieved an accuracy of 71.0%, a sensitivity of 70.0%, a specificity of 71.2%, and an AUC of 0.71 in external validation, which also outperformed other models (Table 3).DISCUSSION and CONCLUSION
In this paper, we proposed a novel OAP-EL model using quantitative sMRI data obtained at term-equivalent age to predict later cognitive deficits at 2 years of age. The results showed that the ensemble learning model with guidance of ontologies was able to outperform multiple traditional machine learning and peer ensemble learning models, demonstrating the feasibility of enhancing EL model performance by incorporating domain knowledge.Acknowledgements
This work was supported by the National Institutes of Health [R01-EB029944, R01-EB030582, R01-NS094200 and R01-NS096037]; The funders played no role in the design, analysis, or presentation of the findings.References
1. Fleiss, B., P. Gressens, and H.B. Stolp, Cortical gray matter injury in encephalopathy of prematurity: link to neurodevelopmental disorders. Frontiers in Neurology, 2020. 11: p. 575.
2. Inder, T.E., et al., Abnormal cerebral structure is present at term in premature infants. Pediatrics, 2005. 115(2): p. 286-294.
3. Rathbone, R., et al., Perinatal cortical growth and childhood neurocognitive abilities. Neurology, 2011. 77(16): p. 1510-1517.
4. Steinman, K.J., et al., Neonatal watershed brain injury on magnetic resonance imaging correlates with verbal IQ at 4 years. Pediatrics, 2009. 123(3): p. 1025-1030.
5. Dong, X., et al., A survey on ensemble learning. Frontiers of Computer Science, 2020. 14(2): p. 241-258.
6. Huang, F., G. Xie, and R. Xiao. Research on ensemble learning. in 2009 International Conference on Artificial Intelligence and Computational Intelligence. 2009. IEEE.
7. Khosla, M., et al., Ensemble learning with 3D convolutional neural networks for functional connectome-based prediction. NeuroImage, 2019. 199: p. 651-662.
8. Lella, E., et al., An Ensemble Learning Approach Based on Diffusion Tensor Imaging Measures for Alzheimer’s Disease Classification. Electronics, 2021. 10(3): p. 249.
9. Liu, J., et al., Multi-view ensemble learning for dementia diagnosis from neuroimaging: An artificial neural network approach. Neurocomputing, 2016. 195: p. 112-116.
10. Zhang, S.-B. and Q.-R. Tang, Protein–protein interaction inference based on semantic similarity of gene ontology terms. Journal of theoretical biology, 2016. 401: p. 30-37.
11. Shen, Y., et al., An ontology-driven clinical decision support system (IDDAP) for infectious disease diagnosis and antibiotic prescription. Artificial intelligence in medicine, 2018. 86: p. 20-32.
12. Köhler, S., et al., Clinical diagnostics in human genetics with semantic similarity searches in ontologies. The American Journal of Human Genetics, 2009. 85(4): p. 457-464.
13. Makropoulos, A., et al., The developing human connectome project: A minimal processing pipeline for neonatal cortical surface reconstruction. Neuroimage, 2018. 173: p. 88-112.
14. Makropoulos, A., et al., Automatic whole brain MRI segmentation of the developing neonatal brain. IEEE transactions on medical imaging, 2014. 33(9): p. 1818-1831.
15. Chen, T. and C. Guestrin. Xgboost: A scalable tree boosting system. in Proceedings of the 22nd acm sigkdd international conference on knowledge discovery and data mining. 2016.
16. Tustison, N. J., Avants, B. B., Cook, P. A., Zheng, Y., Egan, A., Yushkevich, P. A., & Gee, J. C. (2010). N4ITK: improved N3 bias correction. IEEE transactions on medical imaging, 29(6), 1310-1320.
17. Smith, S. M. (2002). Fast robust automated brain extraction. Human brain mapping, 17(3), 143-155.
18. Makropoulos, A., Gousias, I. S., Ledig, C., Aljabar, P., Serag, A., Hajnal, J. V., ... & Rueckert, D. (2014). Automatic whole brain MRI segmentation of the developing neonatal brain. IEEE transactions on medical imaging, 33(9), 1818-1831.
19. Gousias, I. S., Edwards, A. D., Rutherford, M. A., Counsell, S. J., Hajnal, J. V., Rueckert, D., & Hammers, A. (2012). Magnetic resonance imaging of the newborn brain: manual segmentation of labelled atlases in term-born and preterm infants. Neuroimage, 62(3), 1499-1509.
20. Schuh, A., Makropoulos, A., Wright, R., Robinson, E. C., Tusor, N., Steinweg, J., ... & Rueckert, D. (2017, April). A deformable model for the reconstruction of the neonatal cortex. In 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017) (pp. 800-803). IEEE.
Figures
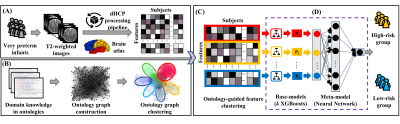
Fig. 1. Schematic diagram of OAP-EL for early prediction of cognitive deficits at 2-year corrected age using brain geometric features derived from T2-weighted MRI data acquired at term equivalent age in very preterm infants. (A) Brain geometric feature extraction, (B) Ontology graph construction and clustering, (C) Ontology-guided feature clustering, and (D) Ensemble learning model. OAP-EL: Ontology-guided Attribute Partitioning Ensemble Learning.
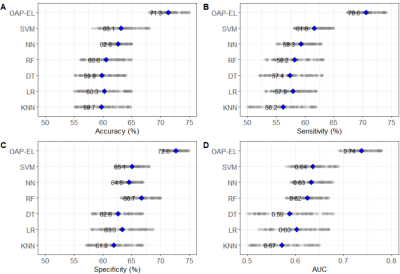
Fig. 2. Internal validation and comparison of our proposed OAP-EL model versus traditional machine learning models on (A) Accuracy, (B) Sensitivity, (C) Specificity, and (D) AUC in early prediction of cognitive deficits. Gray dots indicate metrics of repeated experiments, and blue diamonds indicate mean values. KNN: K-nearest neighbor; LR: Logistic Regression; SVM: Support Vector Machine; DT: Decision Tree; RF: Random Forest; NN: Neural Network; OAP-EL: Ontology-guided Attribute Partitioning Ensemble Learning.
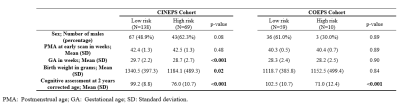
Table 1. Demographics of all very preterm infants at term equivalent age and assessment at 2 years corrected age
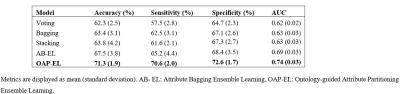
Table 2. Internal validation and comparison of our proposed OAP-EL model versus peer ensemble learning models in early prediction of cognitive deficits.
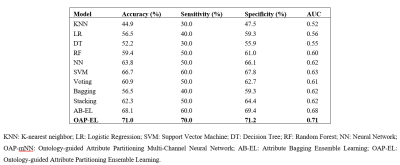
Table 3. External validation and comparison of our proposed OAP-EL model versus traditional machine learning and peer ensemble learning models in early prediction of cognitive deficits.