3519
Undersampling artifact reduction for free-breathing 3D stack-of-radial MRI based on a deep adversarial learning network1Department of Radiological Sciences, University of California Los Angeles, Los Angeles, CA, United States, 2Department of Physics and Biology in Medicine, University of California Los Angeles, Los Angeles, CA, United States, 3Department of Bioengineering, University of California Los Angeles, Los Angeles, CA, United States, 4MR Application Predevelopment, Siemens Healthcare GmbH, Erlangen, Germany, 5MR R&D Collaborations, Siemens Medical Solutions USA, Inc., Cary, NC, United States, 6MR R&D Collaborations, Siemens Medical Solutions USA, Inc., Los Angeles, CA, United States
Synopsis
Undersampling is desired to reduce scan time but can cause streaking artifacts in stack-of-radial imaging. State-of-the-art deep neural networks such as the U-Net can be trained in a supervised manner to remove streaking artifacts but produce blurred images and loss of image details. Therefore, we developed and trained a 3D generative adversarial network to preserve perceptual image sharpness while removing streaking artifacts. The network used a combination of adversarial loss, L2 loss and structural similarity index loss. We demonstrated the feasibility of the proposed network for removing streaking artifacts and preserving perceptual image sharpness.
INTRODUCTION
Undersampled stack-of-radial trajectories allow free-breathing scans but provide noisy images with streaking artifacts. Previous work on deep neural networks used supervised learning to remove streaking artifacts.1-6 Those studies use only pixel-wise loss functions to constrain the output of the network, which leads to image blurring in the output.7,8 Therefore, we propose to use a generative adversarial network (GAN) to preserve perceptual image sharpness while removing streaking artifacts.METHODS
Data Acquisition and PreparationThe study was HIPAA compliant and approved by the institutional review board. A prototypical free-breathing 3D golden-angle stack-of-radial gradient echo sequence with single-echo and multi-echo (6 echoes) options was used.9-12 Written informed consent was obtained for each subject. Data were collected from two institutes: dataset A: 17 healthy subjects at institute 1, dataset B: 3 patients with non-alcoholic fatty liver disease at institute 1 (all on MAGNETOM Prismafit except one healthy subject on MAGNETOM Skyra, Siemens Healthcare, Erlangen, Germany); dataset C: 4 healthy subjects (2-echo images) on a 3T MR-PET scanner (Biograph mMR, Siemens Healthcare, Erlangen, Germany) at institute 2.
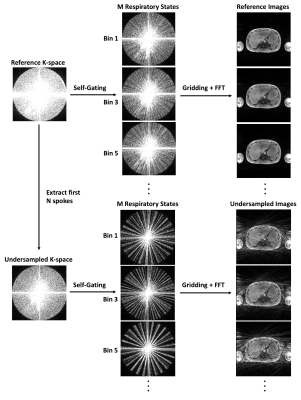
Figure 1 shows the retrospective undersampling and reconstruction framework. Undersampling was achieved by extracting the first N spokes from the reference scan. The k-space data was motion-gated using self-gating followed by image reconstruction using gridding.10,12-14 Dataset A was divided into training (111 sets of 3D images from 8 subjects), validation (29 sets of 3D images from 2 subjects), and testing (91 sets of 3D images from 7 subjects) subsets. Datasets B and C were completely used as the testing data (36 sets of 3D images). Data augmentation of the training and validation data was implemented by self-gating to 5 respiratory states, each fed into the network independently. For testing, respiratory gating with 40% acceptance rate was used to achieve a good balance of motion artifact and scan efficiency. There were 555 pairs of 3D images for training, 145 pairs for validation and 127 pairs for testing. The network was trained on 5x accelerated images and tested on 3x, 4x and 5x accelerated images.
Network Architecture and Training
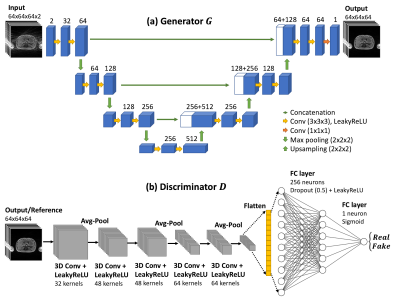
The proposed GAN consisted of a generator and a discriminator. As shown in Figure 2, the generator was adapted from a 3D U-Net, consisting of contracting and expanding paths and skip connections. The discriminator consisted of 5 convolution layers interleaved with 3 average pooling layers, followed by 2 fully-connected layers. The loss function of the network consisted of three parts: $$$L_{total}=\alpha L_G+\beta L_{l_2}+\gamma L_{SSIM}$$$, where $$$L_G$$$ is adversarial loss, $$$L_{l_2}$$$ is L2 loss, and $$$L_{SSIM}$$$ is SSIM loss, the weights for each loss were $$$[\alpha, \beta, \gamma]=[0.6, 0.2, 0.2]$$$.
The input was complex images with real and imaginary parts as two channels and the output was magnitude images. Training and validation data were randomly cropped to 64x64x64 patches and cropping was not used when testing data. The network was trained with 100 epochs using an Adam algorithm15 with β = 0.9. A mini-batch training was performed with 16 batches per iteration. Initial learning rates were 0.0001 for the generator and 0.00001 for the discriminator, respectively.
Implementation and training of the GAN were completed in Python version 3.5 using the Pytorch library version 1.4.0.16 The training and testing were performed on a commercially available graphics processing unit (NVIDIA Titan RTX, 24GB memory). The total training time was about 72 hours.
Performance Evaluation
The developed GAN network was compared to a 3D U-Net. The performance of the networks with respect to the input and the reference was evaluated quantitatively by the normalized mean-squared-error (NMSE), SSIM metrics, and ROI analysis, and qualitatively by radiologists’ scores. Three radiologists with an average of 8 years of abdominal image reading experience independently scored each image with a scale of 1-4 based on two criteria: (1) streaking artifacts (1=severe, 2=moderate, 3=mild, 4=minimal) and (2) perceptual sharpness (1=poor, 2=adequate, 3=good, 4=excellent).
RESULTS
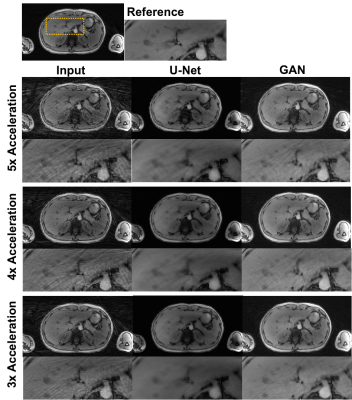
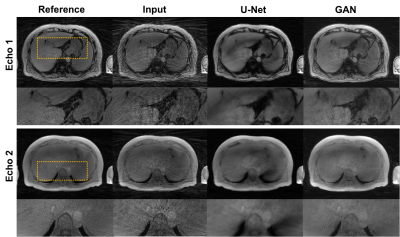
Figures 3 shows an example of a healthy subject from dataset A. Figure 4 shows an example of a patient with 2-echo images. GAN and U-Net successfully reduced streaking artifacts, while GAN appeared sharper than U-Net.Quantitative analysis showed that GAN did not have a significantly different (p<0.068) NMSE than U-Net and both had a significantly lower (p<0.0001) NMSE than the input. GAN had significantly higher SSIM (p<0.003) than U-Net and the input (0.843 vs. 0.794 and 0.738 for 5x acceleration). Quantitative ROI analysis showed that GAN removed the streaking elevated signal and approached the mean signal of the reference images (air: p=0.3497; liver: p=0.1144), whereas U-Net removed the elevated signal only in the air (p=0.3060) and maintained a higher mean in the liver (p=0.0048).
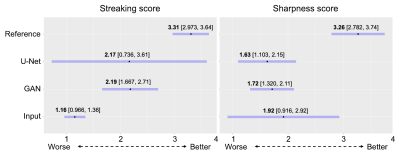
Statistical analysis of radiologists’ scores is shown in Figure 5. Radiologists differed strongly in their opinions of the streaking of the U-Net method, so no comparison with U-Net was significant (p>0.170). Radiologists’ opinions of the GAN method were more consistent, with a 1.02 point improvement in streaking score (p=0.014) without a notable degradation in sharpness compared to the input.
DISCUSSION AND CONCLUSION
A 3D GAN was developed to reduce streaking artifacts and evaluated in subjects. This study showed the feasibility and performance of the proposed network for destreaking. The proposed method can accelerate free-breathing stack-of-radial MRI.Acknowledgements
This study was supported in part by Siemens Medical Solutions USA, Inc and the Department of Radiological Sciences at UCLA.References
1. Hauptmann A, Arridge S, Lucka F, Muthurangu V, Steeden JA. Real‐time cardiovascular MR with spatio‐temporal artifact suppression using deep learning–proof of concept in congenital heart disease. Magn Reson Med. 2019;81(2):1143-1156. doi:10.1002/mrm.27480
2. Nezafat M, El-Rewaidy H, Kucukseymen S, Hauser TH, Fahmy AS. Deep convolution neural networks based artifact suppression in under-sampled radial acquisitions of myocardial T 1 mapping images. Phys Med Biol. 2020;65(22):225024. doi:10.1088/1361-6560/abc04f
3. Fan L, Shen D, Haji‐Valizadeh H, et al. Rapid dealiasing of undersampled, non‐Cartesian cardiac perfusion images using U‐net. NMR in Biomedicine. 2020;33(5). doi:10.1002/nbm.4239
4. Shen D, Ghosh S, Haji‐Valizadeh H, et al. Rapid reconstruction of highly undersampled, non‐Cartesian real‐time cine k ‐space data using a perceptual complex neural network (PCNN). NMR in Biomedicine. 2021;34(1). doi:10.1002/nbm.4405
5. Kofler A, Dewey M, Schaeffter T, Wald C, Kolbitsch C. Spatio-Temporal Deep Learning-Based Undersampling Artefact Reduction for 2D Radial Cine MRI With Limited Training Data. IEEE Trans Med Imaging. 2020;39(3):703-717. doi:10.1109/TMI.2019.2930318
6. Chen D, Schaeffter T, Kolbitsch C, Kofler A. Ground-truth-free deep learning for artefacts reduction in 2D radial cardiac cine MRI using a synthetically generated dataset. Phys Med Biol. 2021;66(9):095005. doi:10.1088/1361-6560/abf278
7. Zhao H, Gallo O, Frosio I, Kautz J. Loss Functions for Image Restoration With Neural Networks. IEEE Trans Comput Imaging. 2017;3(1):47-57. doi:10.1109/TCI.2016.2644865
8. Zhu J-Y, Krähenbühl P, Shechtman E, Efros AA. Generative Visual Manipulation on the Natural Image Manifold. arXiv:160903552 [cs]. Published online December 16, 2018. Accessed March 30, 2021. http://arxiv.org/abs/1609.03552
9. Armstrong T, Dregely I, Stemmer A, et al. Free-breathing liver fat quantification using a multiecho 3D stack-of-radial technique: Free-Breathing Radial Liver Fat Quantification. Magn Reson Med. 2018;79(1):370-382. doi:10.1002/mrm.26693
10. Zhong X, Armstrong T, Nickel MD, et al. Effect of respiratory motion on free‐breathing 3D stack‐of‐radial liver relaxometry and improved quantification accuracy using self‐gating. Magn Reson Med. 2020;83(6):1964-1978. doi:10.1002/mrm.28052
11. Armstrong T, Ly KV, Murthy S, et al. Free-breathing quantification of hepatic fat in healthy children and children with nonalcoholic fatty liver disease using a multi-echo 3-D stack-of-radial MRI technique. Pediatr Radiol. 2018;48(7):941-953. doi:10.1007/s00247-018-4127-7
12. Zhong X, Hu HH, Armstrong T, et al. Free‐Breathing Volumetric Liver and Proton Density Fat Fraction Quantification in Pediatric Patients Using Stack‐of‐Radial MRI With Self‐Gating Motion Compensation. J Magn Reson Imaging. 2021;53(1):118-129. doi:10.1002/jmri.27205
13. Grimm R, Block KT, Hutter J, et al. Self-gating Reconstructions of Motion and Perfusion for Free-breathing T1-weighted DCE-MRI of the Thorax Using 3D Stack-of-stars GRE Imaging. In: Proceedings of the 20th Annual Meeting of ISMRM. p. 3814. 2012.
14. Grimm R, Bauer S, Kiefer B, Hornegger J, Block T. Optimal Channel Selection for Respiratory Self-Gating Signals. In: Proeedings of the 21st Annual Meeting of ISMRM. p. 3749. 2013.
15. Kingma DP, Ba J. Adam: A Method for Stochastic Optimization. arXiv:14126980 [cs]. Published online January 29, 2017. Accessed March 15, 2021. http://arxiv.org/abs/1412.6980
16. Paszke A, Gross S, Massa F, et al. PyTorch: An Imperative Style, High-Performance Deep Learning Library. arXiv:191201703 [cs, stat]. Published online December 3, 2019. Accessed March 15, 2021. http://arxiv.org/abs/1912.01703
Figures