3514
Patch-based AUTOMAP image reconstruction of low SNR 1.5 T human brain MR k-space1Department of Radiology, A.A Martinos Center for Biomedical Imaging / MGH, Charlestown, MA, United States, 2Harvard Medical School, Boston, MA, United States, 3Department of Biomedical Engineering, Boston University, Boston, MA, United States, 4GE Healthcare, Bangalore, India, 5Department of Physics, Harvard University, Cambridge, MA, United States
Synopsis
AUTOMAP has proved itself to be robust to noise especially in the low SNR regimes, however due to the fully connected architecture of the input layer, its applicability to large matrix size datasets has been limited. Here we propose a patched-based trained network that enables the reconstruction of larger datasets. Low SNR single-channel volume coil brain images were acquired at 1.5T with different pulse sequences and reconstructed with the trained model. Results obtained show significant denoising potential. An increase in SNR of 1.5-fold as well as an increase in SSIM was also observed.
Introduction
The main limitation of the deep learning approach AUTOMAP - Automated Transform by Manifold Approximation1, for the end-to-end reconstruction of k-space data is that the input layers in the neural network (NN) architecture are fully connected. This directly impairs the versatility of the approach to reconstruct large matrix size MR datasets, as it requires vast amounts of GPU RAM. The practical uses of AUTOMAP have been mainly to improve ultra-low field (< 0.3T) datasets with low SNR and limited to small matrix sizes2. In the pursuit to overcome this memory limitation, we propose a patch-based image reconstruction approach for large datasets. Low SNR human brain data was acquired at 1.5 T with using a single channel volume coil and two different MR sequences – T2-FSE and T2-Flair of matrix size 256 × 206 and 256 × 192 respectively, were reconstructed using the patch-based AUTOMAP image reconstruction. In this abstract we compare the single channel deep learning reconstruction to a multi-channel coil dataset. Image metrics are also computed.Materials and Methods
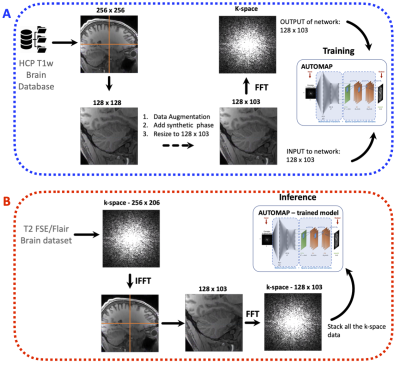
Training set: The training corpus was assembled from 102,000 2D T1-weighted brain MR images selected from the MGH-USC Human Connectome Project (HCP)3 public database. The images were cropped to 256 × 256 and were subsampled equivalently into 4 images of 128 × 128. Synthetic phase modulation was added to the magnitude images to generate complex-valued data. The complex-valued image was then resized to either 128 × 103 or 128 × 96 depending on the data to be reconstructed. To produce the corresponding k-space forward representations for training, each image was Fourier Transformed with MATLAB’s native 2D FFT function and then multiplied by random additive white Gaussian noise (AWGN) ranging from 20 dB to 45 dB as shown in Figure 1a.Architecture of NN: Two networks were trained to learn an optimal feed-forward reconstruction form the k-space domain into the image domain. The real and the imaginary part of datasets were trained together. Both networks were composed of 3 fully connected layers (input layer and 2 hidden layers) of dimension n2 × 1 and activated by the hyperbolic tangent function. The 3rd layer was reshaped to either 128 × 103 or 128 × 96 for convolutional processing. Two convolutional layers convolved 128 filters of 3×3 with stride 1 followed each by a rectifier nonlinearity. The last convolution layer was finally de-convolved into the output layer with 128 filters of 3×3 with stride 1. The output layer formed the complex-valued reconstructed image.
Data Acquisition: Two brain datasets were acquired on two healthy subjects at 1.5T using the single channel volume coil. A T2-FSE MR sequence was acquired with the following parameters: TR= 7000 ms, TE =107 ms, matrix size= 256 × 206, slice thickness = 4.5 mm and 22 slices. A T2-Flair sequence was also acquired with TR= 9000 ms, TE =118 ms, matrix size= 256 × 192, slice thickness = 5 mm and 18 slices. For both MR sequences, a ground truth dataset was also acquired using a multichannel head coil.
Inference Experiment: Each slice of the in vivo raw dataset was first inverse Fourier Transformed into its image domain and divided into 4 quadrant images. Each quadrant image is Fourier Transformed back into its k-space, reconstructed with AUTOMAP (see Figure 1b) and stacked back to form the full image.
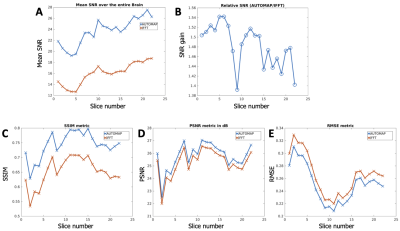
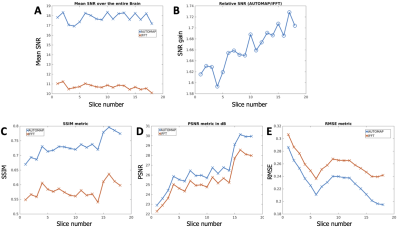
Image Analysis: The signal magnitude of each slice of each dataset was normalized to unity to enable fair comparison between both reconstruction methods. SNR was then computed by dividing the signal magnitude by the standard deviation of the noise. Image quality metrics were evaluated using RMSE (root mean square error), PSNR (Peak Signal-to-Noise Ratio), and SSIM (Structure Similarity Index for Measuring image quality) and the multichannel data was used a reference.
Results
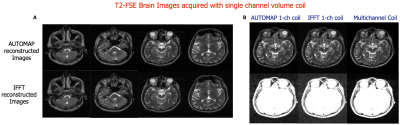
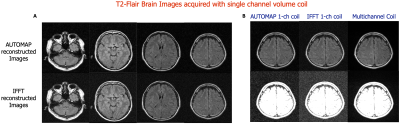
Figures 2 and 4 demonstrate that patch-based AUTOMAP can reconstruct large matrix size datasets from two different MR sequences and as well as improve the image quality. Figures 2B and 4B show the denoising potential of the trained models on large matrix size datasets, where the noise background is significantly reduced with AUTOMAP reconstruction. For the T2-FSE dataset, the results show a 1.4-fold increase in SNR within the brain across all the slices, and we observed an increase in SSIM from 0.6 to 0.8 (see Figure 3). PSNR and RMSE are also improved. The T2-Flair dataset reconstructed with AUTOMAP shows an increase of 1.6-fold in mean SNR within the brain across all the slices. Similar enhancements in SSIM, PSNR and RMSE as the T2-FSE dataset were seen (Figure 5). AUTOMAP reconstruction was able to significantly denoise the one channel volume coil images without any information loss.Discussion and Conclusion
Even if this patch-based trained model is not able to fully take advantage of the sparse low dimensional features present within the full-sized k-space data and its corresponding image, we can still improve the image quality of each patch as well as significantly denoise the background. The next step will be to find additional strategies like adding more convolutional layers to further improve the single channel volume coil images and compare the reconstruction with other deep learning reconstruction approaches.Acknowledgements
We acknowledge support for this work from GE Healthcare, and the National Science Foundation Graduate Research Fellowship under Grant No. DGE-1840990 and the NSF NRT: National Science Foundation Research Traineeship Program (NRT): Understanding the Brain (UtB): Neurophotonics DGE-1633516NSF.References
1.‘Image reconstruction by domain transform manifold learning’, B. Zhu and J. Z. Liu and S. F. Cauley and B. R. Rosen and M. S. Rosen, Nature 555 487 EP - (2018).
2. ‘Boosting the signal-to-noise of low-field MRI with deep learning image reconstruction’, N. Koonjoo, N., B. Zhu, G.C. Bagnall, et al. Sci Rep 11, 8248 (2021).
3. ‘MGH–USC Human Connectome Project datasets with ultra-high b-value diffusion MRI’, Fan, Q. et al. NeuroImage 124, 1108–1114 (2016).
Figures