3508
Volumetric patch-based image reconstruction for enhancing hyperpolarized 13C MRI with hybrid image guidance1Advanced Imaging Research Center, UT Southwestern Medical Center, Dallas, TX, United States, 2Department of Radiology, UT Southwestern Medical Center, Dallas, TX, United States, 3Electrical and Computer Engineering, UT Dallas, Richardson, TX, United States
Synopsis
This study presents a volumetric extension of the previously proposed patch-based algorithm for enhancing spatial resolution of 13C MRI. Feasibility of integrating structural and perfusion images as guidance for the patch-based reconstruction is also demonstrated with a digital phantom. The performance of the method was further tested for a hyperpolarized 13C lactate brain image using hybrid probability maps, created from T1-weighted 1H MRI and perfusion images.
INTRODUCTION
13C MRI with dissolution dynamic nuclear polarization (DNP) is an emerging non-invasive imaging method for investigating in vivo metabolism. In particular, hyperpolarized (HP) [1-13C]pyruvate has been utilized to detect altered carbohydrate metabolism (1). Due to several technical challenges of DNP such as large point spread function and limited signal sensitivities, spatial resolution of HP images is often compromised, hampering precise localization of regions with aberrant metabolism. Over the last couple of years, researchers proposed to exploit structural information that is available from 1H MRI for enhancing spatial resolution of HP 13C images (2-5). The current study extends our previous work that focused on two-dimensional patch-based algorithm to a volumetric reconstruction. In addition, the volumetric patch-based algorithm was applied to include hybrid information such as structural and perfusion as guidance to properly describe 13C molecular distribution patterns.METHODS
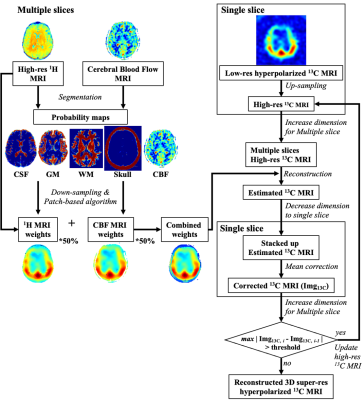
Volumetric patch-based algorithmThe current study proposed a volumetric patch-based method that is composed of three reconstruction steps: a) compartmental segmentation of 1H T1 FLAIR images, b) preparation of initial high-resolution multi-slice 13C images by interpolation, c) iterative reconstruction of super-resolution 13C images using patch-based weighting matrices. The proposed reconstruction scheme is summarized in figure 1.
Phantom simulation
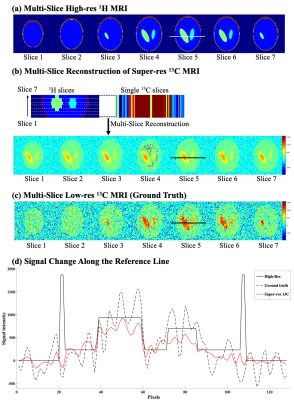
A 3D digital phantom with three compartments was generated using MATLAB (MathWorks, Natick, MA) to evaluate the performance of the proposed algorithm including the in-plane SNR among different segments and through-plane signal reconstruction. Seven slices were generated for 1H MRI (simulated slice thickness = 5 mm, matrix size = 256x256). Two sets of 13C images were simulated for different slice thicknesses, including thin, multiple-slice image set as ground truth images (simulated slice thickness =5 mm, matrix size = 64x64, number of slices = 7), and a single-slice low-resolution image (simulated slice thickness = 30 mm, matrix size = 16x16).
Brain imaging protocol
All MR images were acquired from a clinical 3T 750w wide-bore MRI scanner (GE Healthcare) and a 1H/13C dual-frequency head RF coil (Clinical MR Solutions) (6). The brain imaging protocol included a series of 1H and 13C scans, including a 2D axial T1-weighted 1H FLAIR (TE/TR =12/700 ms, FOV = 24x24 cm2, matrix size = 256x256, slice thickness = 5 mm, number of slices = 13), a 3D 1H pseudo-continuous arterial spin labeling (ASL; TE/TR/TI =9.9/4931/2025 ms, FOV = 24x24x6.5 cm3, matrix size = 128x128x21), and a 2D 13C dynamic spiral imaging (single 3-cm slice, FOV = 24x24 cm2, matrix size = 64x64). For the 13C MRI, the subject (35 y.o. male) was injected with a bolus of HP [1-13C]pyruvate, which was polarized using a SPINlabTM system (GE Healthcare) (7).
Image reconstruction with hybrid image guidance
First, the acquired 1H T1 FLAIR images were segmented using Statistical Parametric Mapping analytic package (SPM12, The Wellcome Centre for Human Neuroimaging, University College London) to generate four probability maps of gray matter, white matter, cerebrospinal fluid and skull. In parallel, the CBF probability map was created by normalization with maximum signal intensity. Then, patch-based weighting matrices were calculated from 7x7x7 neighborhood voxels of the T1 FLAIR and CBF images. The combined weighting matrix and upsampled multi-slice 13C images were iteratively estimated and updated with a mean correction step, which minimizes the signal difference between adjacent iterations. The predicted iteration number to create the final HP 13C MRI was calculated from a power curve fitting function from the first six iterations.
RESULTS
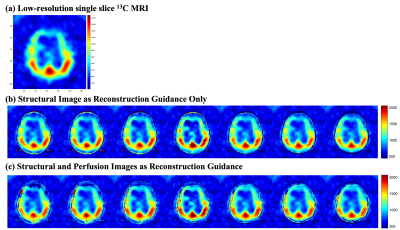
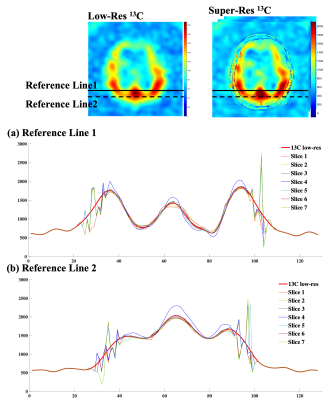
The digital phantom was used to simulate a set of multiple slices high-resolution 1H MRI (figure 2a), low-resolution 13C MRI (figure 2c) and a single slice 13C MRI. The reconstructed multiple slices super-resolution 13C MRI (figure 2b) showed improved in-plane spatial resolution and restored multiple slices information based on the high-resolution structural images. Compared to the low-resolution ground truth (figure 2c), the reconstructed signals were more stable within compartments and preserved the inter-compartmental contrasts as described by the cross-sectional profiles along the reference lines (figure 2d). Compared to the low-resolution ground truth image, non-existed compartments signals appeared in some of the reconstructed images, which might attribute to the lack of information from the high-resolution image patch between slices. The simulation results from the digital phantom demonstrated the feasibility of the proposed three-dimensional patch-based super-resolution algorithm.For the HP 13C brain images, HP [1-13C]lactate image (figure 3a) was used. Overall, the patch-based algorithm with high-resolution structural 1H MRI guidance sharpened the 13C image to enhance the brain structure and the boundary, and reconstructed with through-plane signal distribution (figure 3b). When both structural and CBF images were used, further provide information on the edge of head (figure 3c), instated of the truncated signal distribution on the edge of head while only using the structural image as guidance (figure 3b). While the cross-sectional profiles along the reference lines were maintained within the brain, additional features were observed between the reconstructed image slices (figure 4a, b). Compared with the previous report (5), including perfusion information as an additional guidance image restored truncated 13C signals on edge of head.
CONCLUSION
The current study proposed a patch-based reconstruction method that enhances both in-plane and through-plane resolutions. We showed the feasibility of enhancing image contrasts of low-resolution 13C images using the method.Acknowledgements
National Institutes of Health of the United States (P41 EB015908, S10 RR029119, S10 OD018468, R01 NS107409); The Welch Foundation (I-2009-20190330).References
1. Golman K, Zandt RI, Lerche M, Pehrson R, Ardenkjaer-Larsen JH. Metabolic imaging by hyperpolarized 13C magnetic resonance imaging for in vivo tumor diagnosis. Cancer Res. 2006;66(22):10855-60.
2. Ehrhardt MJ, Gallagher FA, McLean MA, Schonlieb CB. Enhancing the spatial resolution of hyperpolarized carbon-13 MRI of human brain metabolism using structure guidance. Magn Reson Med. 2021.
3. Dwork N, Gordon JW, Tang S, O'Connor D, Hansen ESS, Laustsen C, et al. Di-chromatic interpolation of magnetic resonance metabolic images. MAGMA. 2021;34(1):57-72.
4. Farkash G, Markovic S, Novakovic M, Frydman L. Enhanced hyperpolarized chemical shift imaging based on a priori segmented information. Magn Reson Med. 2019;81(5):3080-93.
5. Ma J, Park JM. Super-Resolution Hyperpolarized (13)C Imaging of Human Brain Using Patch-Based Algorithm. Tomography. 2020;6(4):343-55.
6. Ma J, Pinho MC, Harrison CE, Chen J, Sun C, Hackett EP, et al. Dynamic (13) C MR spectroscopy as an alternative to imaging for assessing cerebral metabolism using hyperpolarized pyruvate in humans. Magn Reson Med. 2021.
7. Nelson SJ, Kurhanewicz J, Vigneron DB, Larson PE, Harzstark AL, Ferrone M, et al. Metabolic imaging of patients with prostate cancer using hyperpolarized [1-(1)(3)C]pyruvate. Sci Transl Med. 2013;5(198):198ra08.
Figures