3489
MAP-MRI diffusion estimates are biased by simultaneous multi-slice acceleration1Neurosurgery, Medical College of Wisconsin, Milwaukee, WI, United States, 2Radiology, Medical College of Wisconsin, Milwaukee, WI, United States, 3GE Healthcare, New York, NY, United States, 4GE Healthcare, Waukesha, WI, United States, 5GE Healthcare, Menlo Park, CA, United States
Synopsis
Advanced diffusion MRI models are being explored to study the complex microstructure of the brain with higher accuracy. However, these techniques require long acquisition times. Simultaneous multi-slice (SMS) accelerates data acquisition by exciting multiple image slices simultaneously and separating the overlapping slices using a mathematical model. However, SMS acceleration introduces increased noise in reconstructed images and crosstalk between simultaneously excited slices. These compounded effects from SMS acceleration could affect tissue microstructure parameters derived from advanced diffusion MRI models.
INTRODUCTION
Diffusion MRI became a versatile tool to study brain white matter. While diffusion tensor imaging (DTI) is the most commonly used technique, its simplified Gaussian diffusion approximation compromises its accuracy. Advanced diffusion MRI models are being explored to study the microstructure of the brain with higher accuracy. However, these techniques typically require longer acquisition times compared to DTI.Simultaneous multi-slice (SMS) acceleration is introduced recently to shorten data acquisition times1–3. SMS excites multiple slices simultaneously and separates the overlapping slices using a mathematical model. However, SMS causes increased noise in reconstructed images that stem from the noise in the measured data and its propagation to the kernel estimation. Additionally, there is residual crosstalk between simultaneously excited slices. As a result, diffusion attenuation measurements in a voxel are affected by the increased noise and residues from the overlapped slices.
In this study, we investigated the bias introduced by SMS acceleration on the estimation of zero-displacement (ZD) and non-Gaussianity (NG) maps obtained from the Mean Apparent Propagator (MAP-MRI) technique4. Two alternative SMS reconstruction techniques are compared to determine if the measurement bias could be reduced by more effective reconstruction.
METHODS
Five healthy volunteers were scanned in a 3T GE Premier scanner. The study was approved by the IRB and participants signed written consents. MAP-MRI data were acquired using four b-values between 1000s/mm2 and 4000s/mm2 and 94 diffusion encoding directions and four b=0 images. A single-shot Spin-Echo EPI with blipped-CAIPI scheme2 was run with 2mm isotropic resolution, TE=84ms and TR=4600ms. To improve reliability and statistical power, two singleband and two 3-band SMS data were acquired from each subject. Shorter TR afforded by the SMS was not utilized to keep all imaging parameters matched between scans, except for the SMS factor. Images were reconstructed using a hybrid-space coil-by-coil slice unaliasing approach called slice-ARC. The general formalism for ARC was published earlier for in-plane acceleration5. Slice-ARC uses a similar approach, calculating kernels from a singleband reference scan. Reconstruction kernels for 3-band SMS were calculated using both slice-ARC method, and split-slice-ARC method, which is a variant of slice ARC that aims to reduce slice leakage, in a manner similar to that presented by Cauley et al6.The outputs of both reconstructions were then corrected for distortion and motion using the TOPUP and EDDY software tools of the FSL7. Repeated scans of each subject were registered using affine transformations. Images were denoised using MP-PCA8 and Gibbs ringing was reduced using DIPY9 tools. MAP-MRI parameter fitting was performed using the DIPY toolbox9 with radial order of 6 and Laplacian regularization. FA maps were also calculated using DTI model and used for registration and analysis.
For group analysis, FA maps from each subject was registered to the standard space FA map using ANTS diffeomorphic registration10. This transformation was then applied to MAP-MRI’s ZD and NG maps. All images were visually inspected to check for artifacts and registration quality. A white matter skeleton was generated for the group with tbss_skeleton script applied on the ANTS registered FA images. An FA skeleton image is then generated by projecting the local FA values onto the skeleton. Then, the skeletonization was applied to the MAP-MRI’s ZD and NG maps. Voxel-based analysis was performed on the skeletonized 4D image series using FSL’s randomise function. The design matrix and contrasts were generated for paired T-test analysis following the GLM example described in FSL wiki page11. Separate comparisons were done between singleband, SMS-3 with slice-ARC or SMS-3 with split-slice-ARC reconstructions. All results were corrected for multiple comparisons and thresholded at p<0.05.
RESULTS
Mean and radial NG estimates were lower in the SMS-3 data compared to the singleband acquisitions (Fig.1). Both slice-ARC and split-slice-ARC resulted in similar biases (left and right panels). Mean and radial NG results were similar, suggesting that the differences were mainly driven by the radial component. Therefore, only mean NG was shown in Fig.1. On the other hand, FA values were higher in the SMS-3 data compared to the singleband (Fig.2). Neither reconstruction method has shown an improvement for FA estimations, either.DISCUSSION
Despite a modest SMS acceleration of 3, there were significantly lower estimates for NG and higher estimates for FA compared to the reference singleband acquisitions. No improvement was observed with either reconstruction method. Although split-slice ARC type approaches were proposed to reduce slice crosstalk, studies reported that this comes at the expense of increased noise6, 12, 13. This could explain increased FA with SMS acceleration since an increase in FA estimates was reported with reduced SNR14.Earlier studies with a single-shell DTI acquisition showed relatively small bias in FA and MD3. This is probably because of the single-shell acquisition, simple Gaussian model of DTI and its muted sensitivity to complex diffusion patterns. However, more advanced diffusion models with multi-shell acquisitions are expected to have higher sensitivity to contamination from increased noise and imperfect slice separation in SMS reconstruction. Our findings are in accord with a recent study that investigated effects of SMS on NODDI15, where they reported significant changes in NODDI parameter maps with SMS acceleration15.
CONCLUSION
Accuracy of diffusion MRI parameter estimates might be compromised when SMS accelerations were used.Acknowledgements
This study is supported in part by GE Healthcare research funds.References
1. Breuer FA, Blaimer M, Heidemann RM, Mueller MF, Griswold MA, Jakob PM. Controlled aliasing in parallel imaging results in higher acceleration (CAIPIRINHA) for multi-slice imaging. Magn Reson Med. 2005; 53: 684-691.
2. Setsompop K, Gagoski BA, Polimeni JR, Witzel T, Wedeen VJ, Wald LL. Blipped-controlled aliasing in parallel imaging for simultaneous multislice echo planar imaging with reduced g-factor penalty. Magn Reson Med. 2012; 67: 1210-1224.
3. Setsompop K, Cohen-Adad J, Gagoski BA et al. Improving diffusion MRI using simultaneous multi-slice echo planar imaging. Neuroimage. 2012; 63: 569-580.
4. Özarslan E, Koay CG, Shepherd TM et al. Mean apparent propagator (MAP) MRI: a novel diffusion imaging method for mapping tissue microstructure. NeuroImage. 2013; 78: 16-32.
5. Brau AC, Beatty PJ, Skare S, Bammer R. Comparison of reconstruction accuracy and efficiency among autocalibrating data-driven parallel imaging methods. Magn Reson Med. 2008; 59: 382-395.
6. Cauley SF, Polimeni JR, Bhat H, Wald LL, Setsompop K. Interslice leakage artifact reduction technique for simultaneous multislice acquisitions. Magn Reson Med. 2014; 72: 93-102.
7. Smith SM, Jenkinson M, Woolrich MW et al. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage. 2004; 23 Suppl 1: S208-19.
8. Veraart J, Fieremans E, Novikov DS. Diffusion MRI noise mapping using random matrix theory. Magn Reson Med. 2016; 76: 1582-1593.
9. Garyfallidis E, Brett M, Amirbekian B et al. Dipy, a library for the analysis of diffusion MRI data. Front Neuroinform. 2014; 8: 8.
10. Avants BB, Tustison NJ, Song G, Cook PA, Klein A, Gee JC. A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage. 2011; 54: 2033-2044.
11. Woolrich MW, Behrens, T. E. J., Beckmann, C. F., Jenkinson, M., Smith, S. M. FSL GLM general advice https://fsl.fmrib.ox.ac.uk/fsl/fslwiki/GLM.
12. Park S, Park J. SMS-HSL: Simultaneous multislice aliasing separation exploiting hankel subspace learning. Magn Reson Med. 2017; 78: 1392-1404.
13. Setsompop K, Cauley SF, Bhat H, Polimeni J, Wald LL. Characterization and mitigation of signal leakage in simultaneous multi-slice (SMS) acquisition. Intl Soc Mag Reson Med. 2013; Proceedings: 3315.
14. Farrell JA, Landman BA, Jones CK et al. Effects of signal-to-noise ratio on the accuracy and reproducibility of diffusion tensor imaging-derived fractional anisotropy, mean diffusivity, and principal eigenvector measurements at 1.5 T. J Magn Reson Imaging. 2007; 26: 756-767.
15. Bouyagoub S, Dowell NG, Gabel M, Cercignani M. Comparing multiband and singleband EPI in NODDI at 3 T: what are the implications for reproducibility and study sample sizes. MAGMA. 2020;
Figures
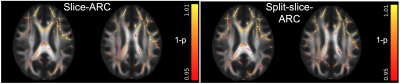
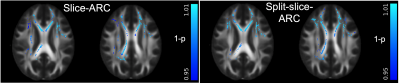
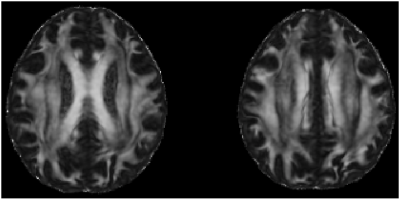
Fig.3. Sample FA maps registered to standard space using ANTS. DTI model is fit using the complete multi-shell diffusion acquisition described in the text.