3467
Contrastive Learning of Inter-domain Similarity for Unsupervised Multi-modality Deformable Registration1Computer Science and Engineering, New York University, New York City, NY, United States, 2Hyperfine, New York City, NY, United States, 3Yale University, New Haven, CT, United States
Synopsis
We propose an unsupervised contrastive representation learning framework for deformable and diffeomorphic multi-modality MR image registration. The proposed deep network and data-driven objective function yield improved registration performance in terms of anatomical volume overlap over several previous hand-crafted objectives such as Mutual Information and others. For fair comparison, our experiments train all methods over the entire range of a key registration hyperparameter controlling deformation smoothness using conditional registration hypernetworks. T1w and T2w brain MRI registration improvements are presented across a large cohort of 1041 high-field 3T research-grade acquisitions while maintaining comparable deformation smoothness and invertibility characteristics to previous methods.
Introduction
Inter-modality deformable image registration is crucial to several clinical and research workflows, including motion correction, spatial normalization to templates for segmentation and standardized statistics, and intra-operative registration of portable imaging to clinical scans. While several works have tackled inter-modality image registration1,2,3,4, they perform considerably worse than methods working in the intra-modality setting due to the difficulty of reliably estimating anatomical similarity across imaging domains.Most commonly used inter-modality MRI similarity functions include information-theoretic approaches working on intensity histograms (e.g. mutual information and its local extensions1,2]), approaches focusing on edge alignment (e.g. normalized gradient fields3), and those that build local descriptors invariant to imaging domains (e.g. modality invariant neighborhood descriptors4). However, due to their hand-crafted nature, these functions typically require significant domain expertise, non-trivial tuning, and may not consistently generalize outside of the domain-pair that they were originally proposed for.
In this work, we propose a fully unsupervised and data-driven deep network approach to learning an inter-domain similarity function that is sensitive to deformations across multiple scales and used for deformable image registration. Once registered, our approach ensures that corresponding locations have high mutual information in a deep data-driven representation space. Our approach extends recent developments in Patch-based Noise Contrastive Estimation5 (PatchNCE) to inter-subject registration by using a pretrained feature space. The proposed method is demonstrated to yield improved registration with higher anatomical overlap as compared to several previous baseline similarity functions.
Materials and Methods
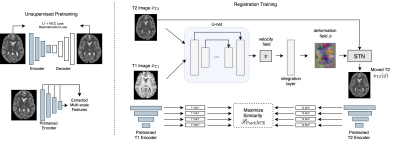
Imaging: Processed 3T T1w and T2w scans and anatomical segmentations are acquired from the S1200 HCP6 data release and are downsampled to 2 x 2 x 2 mm3 resolution for rapid prototyping. We use 800/50/191 subjects as training, validation, and testing data for inter-subject registration, respectively.Training: The overall training pipeline is illustrated in Fig. 1. We align images by maximizing their deep feature similarity and train in two stages.
In stage 1, we train two domain-specific autoencoders with a joint $$$L_1$$$ + Local Normalized Cross Correlation reconstruction objective on their respective training sets. Once trained, their encoders are frozen and used as domain-specific multi-scale feature extractors in stage 2.
In stage 2, a deterministic VoxelMorph registration network7 takes a pair of images $$$x_{T1}$$$ and $$$x_{T2}$$$ to be registered as input and yields a stationary velocity field (SVF) $$$v$$$ between $$$x_{T1,T2}$$$ which can be efficiently integrated to obtain a dense displacement field.
Once warped, features are extracted from the (target image, moved image)-pairs by their frozen domain-specific encoders from stage 1. These features are then projected onto a representation space by trainable multilayer perceptrons (as in SimCLR8) where the similarity between multiscale features at corresponding locations from the target and moved images can be maximized (as in PatchNCE5). We further use a global mutual information loss to maintain global alignment for a final registration loss of (0.1 PatchNCE + 0.9 MI), abbreviated to (NCE + MI) in the remainder of this abstract. To ensure smooth deformations, we employ a diffusion regularizer $$$\|v\|_{2}^2$$$ on $$$v$$$.
Importantly, as all methods are sensitive to regularization strength9, we use a hypernetwork conditioned on diffusion regularization strength to sweep over all possible regularization values as in Mok, et al.10 As a result, all hypernetwork registration models were trained with $$$(1 - \lambda)L_{reg}(x_{T1}, \phi \circ x_{T2}) + \lambda \|v\|_{2}^2$$$ as a loss function where $$$\lambda$$$ is a hyperparameter uniformly sampled from [0, 1] during training and $$$L_{reg}$$$ represents the various similarity functions to be benchmarked.
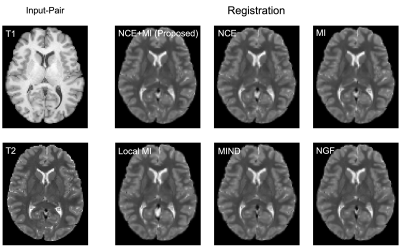
Evaluation: We benchmark all methods using the commonly used inter-subject registration task7,9,10 and report Dice overlap between the segmentation of the target image and the moved segmentation of the moving image, indicating registration correctness; and the percentage of folding voxels, indicating deformation quality and invertibility. The proposed loss function was compared against MI1, Local MI2, NGF3, and MIND4 as baselines while maintaining the same network architecture. At testing time, we sweep over the range of $$$\lambda$$$ in increments of 0.1 thus generating several registration networks for each benchmarked loss function.
Results
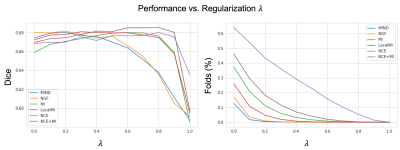
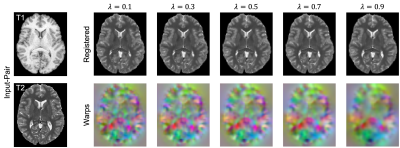
Quantitatively, on a held-out test set of 191 subjects, (Fig 2, left) we see that our method (NCE+MI) with $$$\lambda$$$ = 0.6 - 0.8 outperforms all other losses over all of their respective $$$\lambda$$$’s in terms of anatomical/Dice overlap. Further, (Fig. 2, right) our method (NCE+MI) achieves high registration accuracy while maintaining a comparably negligible percentage of folding voxels to other methods in the $$$\lambda$$$ > 0.6 range, simultaneously indicating high registration accuracy alongside smooth and diffeomorphic deformations. In Fig. 3, we see that the proposed loss function yields more accurate registration perceptually. In Fig. 4, we show that the hypernetwork model generates the expected warped images and deformation fields, with low $$$\lambda$$$’s yielding strong deformations and high $$$\lambda$$$’s yielding highly regular deformations.Discussion
We present an unsupervised contrastive learning framework for multi-modality diffeomorphic registration. Our results demonstrate improved registration accuracy on a large public database while maintaining smooth and invertible deformations. Experimentally, we validate our claims by benchmarking against several previous hand-crafted loss functions and automatically tune deformation regularity for optimal performance for all methods considered by using hypernetworks. Future work will include extensions of our methodology to other use cases such as high-field to low-field MRI registration and intra-operative multi-modality registration.Acknowledgements
No acknowledgement found.References
- Wells III, William M., et al. "Multi-modal volume registration by maximization of mutual information." Medical image analysis 1.1 (1996): 35-51.
- Guo, Courtney K. Multi-modal image registration with unsupervised deep learning. Diss. Massachusetts Institute of Technology, 2019.
- Haber, Eldad, and Jan Modersitzki. "Intensity gradient based registration and fusion of multi-modal images." International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, Berlin, Heidelberg, 2006.
- Heinrich, Mattias P., et al. "MIND: Modality independent neighbourhood descriptor for multi-modal deformable registration." Medical image analysis 16.7 (2012): 1423-1435.
- Park, Taesung, et al. "Contrastive learning for unpaired image-to-image translation." European Conference on Computer Vision. Springer, Cham, 2020.
- Van Essen, D.C., et al: The WU-Minn human connectome project: an overview. Neuroimage 80, 62-79 (2013).
- Dalca, Adrian V., et al. "Unsupervised learning of probabilistic diffeomorphic registration for images and surfaces." Medical image analysis 57 (2019): 226-236.
- Chen, Ting, et al. "A simple framework for contrastive learning of visual representations." International conference on machine learning. PMLR, 2020.
- Hoopes, Andrew, et al. "Hypermorph: Amortized hyperparameter learning for image registration." International Conference on Information Processing in Medical Imaging. Springer, Cham, 2021.
- Mok, Tony CW, and Albert Chung. "Conditional Deformable Image Registration with Convolutional Neural Network." International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, Cham, 2021.
Figures