3465
Predicting Breath-Hold Liver Diffusion MRI from Free-Breathing Data using a Convolutional Neural Network (CNN)
Emmanuelle M. M. Weber1, Xucheng Zhu2, Patrick Koon2, Anja Brau2, Shreyas Vasanawala1, and Jennifer A. McNab1
1Stanford, Stanford, CA, United States, 2GE Healthcare, Menlo Park, CA, United States
1Stanford, Stanford, CA, United States, 2GE Healthcare, Menlo Park, CA, United States
Synopsis
To reduce artifacts in free breathing single-shot diffusion MRI of the liver, UNet based convolutional neural networks were trained to predict breath-hold data from free-breathing data using: 1) simulated data based on a digital phantom and 2) 31 scans of a healthy volunteer. The developed networks successfully reduced motion induced artifacts in DWI images.
Introduction
Abdominal single-shot diffusion-weighted spin echo EPI (DW-SE-EPI) is a routine clinical acquisition to detect and characterize small lesions in the liver1,2. Liver diffusion MRI is challenging due to the large field of view and abdominal motion mainly introduced from adjacent organs including the lungs and heart. Motions usually lead to aliasing, signal loss and slice mismatch in the diffusion weighted images (DWI). Therefore, abdominal motion, especially respiratory motion, limit acquisition speed, achievable b-values, resolution and data consistency3-5. Respiratory gating allows breathing phases to be synchronized using either bellows or a navigator-triggered acquisition. Unfortunately, shallow and/or inconsistent breathing or high iron content might limit the navigator's efficacy. Scanning during a breath-hold (BH) can largely reduce the motion but heavily depends on the patient's capacities and is often not feasible for sick patients. In such cases, free-breathing (FB) with multiple averages is used. However, the scan time is generally longer than standard clinical acquisitions and blurring and ghosting artifacts may still degrade the image quality6. Here, a convolution neural network (CNN) based technique is proposed to reduce motion artifacts in liver diffusion MRI by generating the motion free DWI images from free-breathing DWI images.Methods
Digital phantom simulations: Abdominal diffusion-weighted images at different breathing motion phases were simulated for b=50 s/mm2 using an open-source digital phantom7. The motion corrupted images were obtained by averaging the images at different breathing phases. To obtain more realistic data, the amplitude of motion, duration of the respiratory cycle of the phantom were varied for different datasets (A-P: 5 to 10 mm, S-I: 8 to 20 mm R-L: 1 to 8mm, respiration duration: 400 to 5000ms, temporal resolution: 70 ms, number of images per breathing cycle: 19 to 70) as well as the MRI simulation parameters (matrix size : 124x124 to 256x256, TE: 40 or 70ms, TR: 6 or 8s, slice thickness: 3 to 12mm).MRI acquisition: A dataset of 31 paired free-breathing (FB) and end-of-exhalation-breath-hold (BH) axial DWI was acquired on a healthy volunteer in a GE 3T Signa Premier MRI. Single-shot DW-SE EPI were acquired with a range of different scan parameters (trace encoding with two b-values: either 50 and 800s/mm2 or 50 and 1000s/mm2, Nex= 1, 2 or 3 for the low b-value, Nex=4, 6 or 10 for the high-b-value, slice thickness ranging from 3 to 8 mm, in-plane voxel size from 1.5x1.5 to 3x3 mm2, in-plane acceleration factor of 2 or 3, slice spacing between 0 and 1mm, scan time 13 to 30s).
Prediction of motionless DWI: A 16-channel 2D UNet architecture was developed in Keras (Tensorflow) to predict the motionless digital phantom image from the motion corrupted image. The input was first downsampled from 256x256 down to 16x16 during the encoding phase. The encoding phase was composed of four CNN-CNN-pooling blocks, each CNN with a kernel size of 3x3, a stride of 1 and a ReLU activation function, and each 2D MaxPooling layer with a kernel size of 2x2. After the encoding step, the data were upsampled back to their original size during the decoding phase which is composed of 4 blocks of CNN-CNN (kernel size: 3x3, stride: 2x2, activation: ReLU). A similar approach was used to correct motion artifacts on the healthy volunteer dataset except that 3D layers were used instead of 2D layers and each block was composed of three convolutional layers with kernel size of 5x5. In each block, a skip connection and 0.5 dropout regularization layers were used to limit gradient vanishing effects. For both models, the Adam optimizer was used with the mean squared error function and respective 1x10-5 and 1.4x10-4 learning rates for the 2D UNet and 3D UNet. The dataset was separated in 0.6/0.2/0.2 and 0.7/0.15/0.15 batches for training/validation/testing. The model performance during hyperparameters optimization was tracked using the W&B toolbox8.
Results and Discussion
The ghosting generated by averaged repeated free-breathing acquisitions was reduced in both the simulated and healthy volunteer datasets using UNet architectures. After 500 epochs, the model training/validation losses reached 0.001/0.002 and 0.002/0.005 respectively for the phantom and healthy volunteer images. The models’ performances on the test sets were 0.006 and 0.01 error for the 2D and 3D networks respectively. Extension of the network to 3D is more computationally demanding but provides critical information about the significant motion occurring along the SI orientation. While the predicted phantom images were sharp, the in-plane image sharpness on the healthy volunteer dataset was not fully restored. Acquiring additional data and further hyperparameter optimization and increased number of epochs is expected to improve the prediction capabilities. Future work will also test a semi-supervised framework using a conditional Generative Adversarial Network9,10. Nonetheless, the results presented here inspire confidence that a deep learning approach for obtaining high-quality liver diffusion MRI from standard free-breathing diffusion acquisitions is a tractable way to improve clinical image quality.Acknowledgements
No acknowledgement found.References
- Shenoy-Bhangle A. et al, Diffusion weighted magnetic resonance imaging of liver: Principles, clinical applications and recent updates. World J Hepatol., 2017, 9(26): 1081-1091
- Taouli, B. and Koh D.-M., Diffusion-weighted MR Imaging of the Liver, Radiology, 2010, 254(1):47-66
- Kandpal H. et al, Respiratory-Triggered Versus Breath-Hold Diffusion-Weighted MRI of Liver Lesions: Comparison of Image Quality and Apparent Diffusion Coefficient Values, American Journal of Roentgenology, 2009, 192(4): 915-922
- Taouli, B. et al, Diffusion-weighted imaging of the liver: Comparison of navigator triggered and breathhold acquisitions. J. Magn. Reson. Imaging, 2009, 30: 561-568
- Schraml C. et al, Navigator respiratory-triggered diffusion-weighted imaging in the follow-up after hepatic radiofrequency ablation-initial results. J Magn Reson Imaging., 2009, 29: 1308-1316.
- Szklaruk J. et al, Comparison of free breathing and respiratory triggered diffusion-weighted imaging sequences for liver imaging. World J Radiol., 2019, 11(11): 134-143
- Lo W.C. et al, Realistic 4D MRI abdominal phantom for the evaluation and comparison of acquisition and reconstruction techniques. Magn Reson Med., 2019, 81(3): 1863-1875
- Biewald L., “Experiment Tracking with Weights and Biases,” Weights & Biases.
- Johnson, PM and Drangova, M., Conditional generative adversarial network for 3D rigid-body motion correction in MRI. Magn Reson Med., 2019, 82: 901– 910
- Usman, M., Latif, S., Asim, M. et al. Retrospective Motion Correction in Multishot MRI using Generative Adversarial Network. Sci Rep, 2020, 10: 4786
Figures
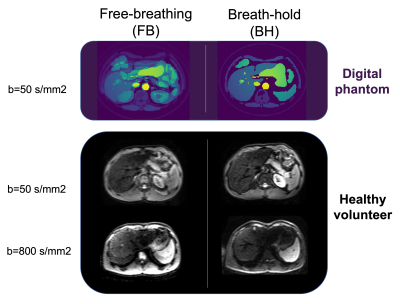
Fig. 1. Digital phantom and healthy volunteer datasets. The free
breathing data (left) contains motion artifacts that do not appear on the
breath hold data set (right). The latter serves as ground truth to train deep
learning models.
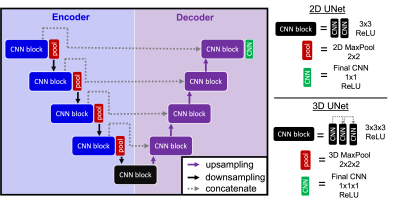
Fig. 2. 2D and 3D deep learning architectures used to reduce
motion artifacts in liver DWI. The 2D UNet was used to reduce motion artifacts
in the phantom whereas the 3D model has been optimized on the healthy volunteer
DWI data.
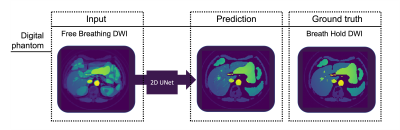
Fig. 3.
Digital phantom DWI (b=50s/mm2) demonstrating the capability of the
2D UNet architecture to predict breath hold quality data from free breathing
data. The developed network efficiently reduced simulated motion artifacts.
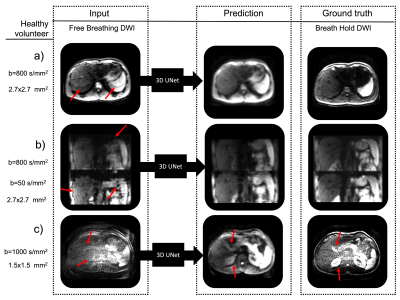
Fig.
4. Healthy
volunteer DWI demonstrating the capability of the 3D UNet architecture to
predict breath hold quality data from free breathing data. The network
efficiently reduced the in-plane and through-plane artifacts. In (a) a
reduction of the aliasing artifacts can be observed (red arrows). In (b), there
is a reduction of slice offsets seen in the sagittal view (red arrows) and in (c)
a reduction of noise and increase in tissue contrast can be observed (red
arrows).
DOI: https://doi.org/10.58530/2022/3465