3453
The impact of streak-removal on deep learning reconstruction of radial datasets1Applied Mathematics, University of Arizona, Tucson, AZ, United States, 2Electrical and Computer Engineering, University of Arizona, Tucson, AZ, United States, 3Department of Medical Imaging, University of Arizona, Tucson, AZ, United States, 4Houston Methodist Hospital, Houston, TX, United States, 5Biomedical Engineering, University of Arizona, Tucson, AZ, United States
Synopsis
Recently, deep learning models have been developed for reconstructing data acquired using radial turbo spin echo sequences, yielding images at multiple echo times as well as co-registered T2 maps. In radial imaging, streaking artifacts from anatomical regions where the magnetic field gradients are nonlinear can obscure pathology and impact accuracy of parameter mapping. In this work, we demonstrate that removal of streaking artifacts from data prior to training can provide substantial improvement in the reconstruction performance of deep learning methods.
Introduction
Radial MRI is naturally robust to motion-induced artifacts1 and has been shown to accelerate scan times in parameter mapping applications2.Using the Radial Turbo-Spin-Echo (RADTSE)3 sequence, data acquired within a single breath hold can be used to create images at multiple echo times (TEs) as well as a co-registered T2 map. In abdominal imaging, it is common to observe streaking artifacts originating from the arms due to gradient non-linearity at the edges of the magnet. Recently, a technique known as CACTUS4 has been shown to successfully remove these artifacts. CACTUS identifies the source of interference within the data and projects the data into the null space of this interference. Importantly, CACTUS can be used in either image domain or in k-space, allowing pre-processing of k-space data before image reconstruction. Multi-scale ResNet (MS-ResNET)5 is a recent deep learning framework that was shown to be successful in replacing iterative reconstructions with fewer artifacts in less time on RADTSE datasets. In this work, we investigate whether using CACTUS pre-processing can improve the reconstruction performance of MS-ResNET on RADTSE datasets.Methods
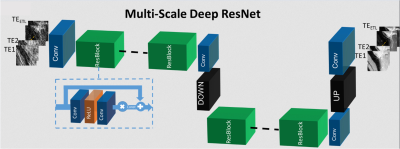
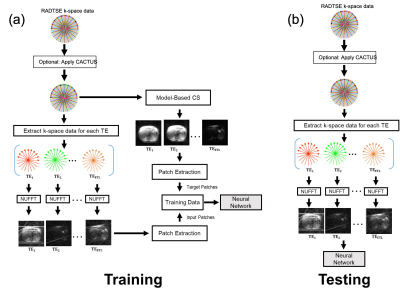
RADTSE abdomen datasets from 21 consenting subjects acquired on a 1.5T Siemens Aera scanner with echo spacing=7.3ms, ETL=32, TR=2500ms, and 6 lines per TE with 256 readout points per line were used in these experiments. 17 datasets were used in training, 2 in validation, and 2 for testing. The data were first reconstructed by applying a non-uniform fast Fourier transform (NUFFT)6 on undersampled k-space data at each TE. This data was used as input to train MS-ResNET. Since it is not practical to acquire fully-sampled RADTSE data in the abdomen, TE images obtained from an iterative model-based compressed sensing method (LLR)7 were created and used as targets. It should be noted that while this model-based technique provides high-quality TE images, it is computationally complex and is not practical for use in a routine clinical setting. 32x32 patches from input and target datasets were generated for training. The overall training and testing procedure is shown in Figure 1. The MS-ResNET consists of convolutional layers with 3x3 kernels and 128 filters. The network architecture is shown in Figure Figure 2. The original RADTSE datasets were then pre-processed using CACTUS to remove streaking artifacts. NUFFT and model-based compressed sensing were applied to the de-streaked k-space data and a new training dataset was created. MS-ResNET was trained again using these input-target pairs.Results and Discussion
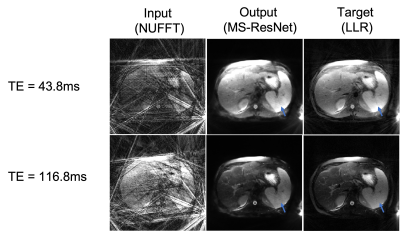
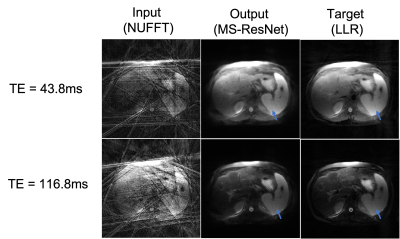
Figure 3 shows images from a test subject at two TEs for the network input, MS-ResNET prediction, and the target LLR technique without CACTUS pre-processing. It can be seen that both the network prediction as well as the target LLR technique suffer from streaking artifacts. Figure 4 shows the same slice reconstructed using different techniques after the original k-space data was pre-processed using CACTUS, using MS-ResNET trained on de-streaked datasets.Our results suggest that removal of streaking artifacts from k-space data using CACTUS prior to generation of the training data can provide substantial improvement in the reconstruction performance of deep learning methods. While we illustrated these improvements using MS-ResNET, we expect these results to hold for other deep learning frameworks.
Conclusion
Using CACTUS pre-processing in k-space to remove streaking artifacts due to magnetic field gradients prior to generation of the training data for deep learning methods can lead to substantially improved reconstruction performance.Acknowledgements
The authors would like to acknowledge grant support from NIH (CA245920), the Arizona Biomedical Research Commission (CTR056039), and the Technology and Research Initiative Fund Technology and Research Initiative Fund (TRIF).References
[1] G.H. Glover and J.M. Pauly. Projection reconstruction techniquesfor reduction of motion effects in MRI.Magnetic resonance in medicine,28(2):275–289, 1992.
[2] M. I. Altbach, T. P. Trouard, and A. F. Gmitro. Radial MRItechniques for obtaining motion-insensitivehigh-resolution images withvariable contrast. pages 125–128. IEEE, IEEE, 2002.
[3] M. I. Altbach et al. Radial fastspin-echo method for T2-weighted imaging and T2 mapping of theliver.Journalof Magnetic Resonance Imaging, 16(2):179–189, 2002.
[4] Zhiyang Fu, Maria Altbach, and Ali Bilgin. Cancellation ofstreak artifacts using the interference null space(CACTUS) for radialabdominal imaging. International Society for Magnetic Resonance inMedicine, ISMRM,2021.
[5] Z. Fu, S. Mandava, Keerthivasan M.B., M.I. Martin, M. Altbach,and A. Bilgin. A Multi-Scale Deep ResNetfor MR Parameter Mapping.International Society for Magnetic Resonance in Medicine, ISMRM, 2018.
[6] Jeffrey A Fessler. http://web.eecs.umich.edu/fessler/irt.2
[7] C. Huang, C.G. Graff, E. Clarkson, A. Bilgin, and M. Altbach. T2 mapping from highly undersampled databy reconstruction of principal component coefficient maps using compressed sensing.Magnetic resonance inmedicine, 67(5):1355–1366, 2012.
Figures