3448
Integrating Subspace Learning, Manifold Learning, and Sparsity Learning to Reconstruct Image Sequences1Department of Electrical and Computer Engineering, University of Illinois at Urbana-Champaign, Urbana, IL, United States, 2Beckman Institute for Advanced Science and Technology, University of Illinois at Urbana-Champaign, Urbana, IL, United States, 3Institute for Medical Imaging Technology, Shanghai Jiao Tong University, Shanghai, China, 4School of Biomedical Engineering, Shanghai Jiao Tong University, Shanghai, China
Synopsis
Many imaging applications, such as dynamic imaging and multi-contrast imaging, involve the acquisition of a sequence of images. This work addresses the underlying image reconstruction problem by incorporating priori information such as partial separability, image sparsity, and manifold structure jointly to enable high-quality image reconstruction from highly sparse data. To this end, we propose a new deep learning-based framework that enforces those constraints effectively and consistently. The proposed method has been validated using multi-contrast imaging data and produced impressive results. The image reconstruction framework can be extended for incorporating additional constraints and/or solving other sequential image reconstruction problems.
Introduction
Reconstruction of image sequences is an essential step in many imaging applications such as dynamic imaging, spectroscopic imaging, and quantitative parameter mapping. A key practical challenge is the lack of sufficient k-space data per frame. To address this problem, significant efforts have been made to use a priori information to compensate for the lack of sufficient experimental data. Commonly exploited image priors include partial separability1, image sparsity2, and manifold structure3,4 and these constraints have been successfully used separately in different reconstruction methods1-8. This work presents a deep learning-based framework for synergistic integration of these image priors through a unified reconstruction formulation. The proposed method has been evaluated using experimental data from multi-contrast imaging, producing significantly improved reconstructions over the existing methods.Methods
Problem FormulationThe problem of reconstructing image sequences can be formulated, in general, as follows. Given a set of sparsely sampled Fourier data points at spatial frequencies $$$\{\boldsymbol{k}_n\}_{n=1}^N$$$ and sequential indexes $$$\{t_m\}_{m=1}^M$$$:
$$\hspace{12em}d\left(\boldsymbol{k}_n,t_m\right)=\int\rho\left(\boldsymbol{x},t_m\right)e^{-i2{\pi}\boldsymbol{k}_{n}{\cdot}\boldsymbol{x}}d\boldsymbol{x}\hspace{12em}(1)$$
reconstruct $$$\rho\left(\boldsymbol{x},t_m\right)$$$. Eq. (1) can be also written in vector form as:
$$\hspace{17.5em}d=F_{\Omega}\rho,\hspace{17.5em}(2)$$
where $$$d\in\mathbb{C}^{M\times{N}}$$$ and $$$\rho\in\mathbb{C}^{M\times{P}}$$$ denote the matrix-form of measured data and desired images, respectively, and $$$F_{\Omega}$$$ represents the physical imaging operator integrating sparse sampling.
In practice, $$$\{\boldsymbol{k}_n\}_{n=1}^N$$$ very sparsely sample $$$k$$$-space for each data frame; so the inverse problem in Eq. (1) is highly under-determined. We solve this problem using a deep learning-based framework that synergistically incorporates subspace learning, manifold learning, and sparsity learning.
Proposed Reconstruction Framework
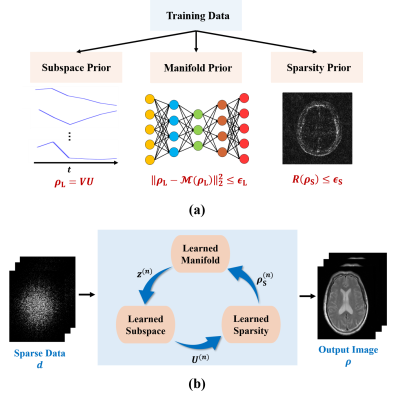
As illustrated in Fig. 1, the proposed reconstruction framework consists of two key components: a) a learning component that learns various image priors from training data, and b) an image reconstruction component that integrates the learned priors.
The learning component captured the following image priors from training data. First, a subspace representation was learned for image sequences exploiting the partial separability1. Specifically, dominant image features were captured and represented by subspace model: $$$\rho_{\text{L}}=VU$$$ with $$$V\in\mathbb{C}^{M\times{R}}$$$ being the subspace bases and $$$U\in\mathbb{C}^{R\times{P}}$$$ the coefficients; the subspace structure was determined from training data through principal component analysis as was done in9. Second, the manifold structure underlying the image sequences in a training set was captured using deep learning. To this end, a deep autoencoder (DAE) was trained under the minimum mean-squared-error principle as was done in4,10. Finally, a sparsity-promoting regularizer was learned to constrain the recovery of the novel image component $$$\rho_{\text{S}}$$$. Particularly, a neural network was trained that unrolled the gradient descent algorithm for solving the following reconstruction problem:
$$\hspace{13.5em}\min_{\rho_{\text{S}}}\left\|d_{\text{S}}-F_{\Omega}\rho_{\text{S}}\right\|_2^2+R(\rho_{\text{S}}).\hspace{13.5em}(3)$$ The resulting unrolled network, as described in10-13, implicitly found an optimal regularization functional $$$R(\cdot)$$$ for recovery of $$$\rho_{\text{S}}$$$.
The image reconstruction component synergistically integrated the learned image priors through a physical model-based constrained reconstruction formulation:
$$\min_{\rho_{\text{L}},\rho_{\text{S}}}\left\|d-F_{\Omega}\left(\rho_{\text{L}}+\rho_{\text{S}}\right)\right\|_2^2\\{\text{subject to:}}\\\hspace{8.5em}{\rho_{\text{L}}=VU},\:\left\|\rho_{\text{L}}-\mathcal{M}\left(\rho_{\text{L}}\right)\right\|_2^2\leq\epsilon_{\text{L}},\:\text{and}\:{R}\left(\rho_{\text{S}}\right)\leq\epsilon_{\text{S}},\hspace{8.5em}(4)$$ where $$$\mathcal{M}$$$ denotes the learned DAE model. The integration of different image priors offers several advantages over the use of a single prior for image reconstruction. First, reconstruction using the subspace representation alone could be ill-conditioned due to sparse sampling, thus leading to significant image artifacts. By integrating the subspace with manifold-based prior, we could effectively constrain the image variations allowed, thus improving the performance. Second, reconstruction incorporating manifold-based prior alone may end up at some poor local minimum due to the highly non-convex nature of image manifold. This problem could be relieved by constraining the solution onto a pre-learned image subspace. Finally, the learned subspace and manifold priors enable better reconstruction of $$$\rho_{\text{L}}$$$; as a result, $$$\rho_{\text{S}}$$$ becomes much sparser, thus making the learned sparsity constraint more effective for image reconstruction.
The constrained optimization problem in Eq. (4) can be solved using the following iterative algorithm:
$$\hspace{1em}\boldsymbol{\textbf{M-step}}:\:z^{(n+1)}=\mathcal{M}\left(VU^{(n)}\right)\hspace{20em}\\\hspace{7.2em}{\boldsymbol{\textbf{U-step}}}:\:U^{(n+1)}=\arg\min_{U}\left\|d_{L}-F_{\Omega}VU\right\|_2^2+\lambda_{L}\left\|VU-z^{(n+1)}\right\|_2^2\hspace{12em}\\\hspace{1.1em}\text{with}\:d_{\text{L}}=d-F_{\Omega}\rho_{\text{S}}^{(n)}\hspace{16em}\\\hspace{1em}{\boldsymbol{\rho_{\text{S}}}\textbf{-step}}:\:\rho_{\text{S}}^{(n+1)}=\mathcal{H}_{\text{S}}\left(d_{\text{S}}\right)\hspace{21.2em}\\\hspace{8.0em}\text{with}\:d_{\text{S}}=d-F_{\Omega}VU^{(n+1)},\hspace{19.5em}(5)$$ where $$$\mathcal{H}_{\text{S}}(\cdot)$$$ is the unrolled network for recovery of $$$\rho_{\text{S}}$$$. After convergence, the final reconstruction was synthesized as: $$$\hat{\rho}=V\hat{U}+\hat{\rho}_{\text{S}}$$$.
Results
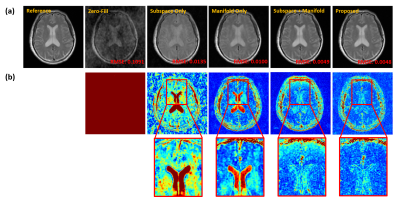
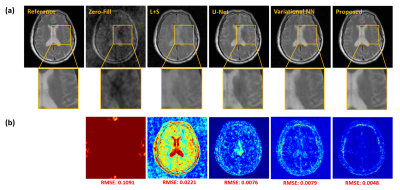
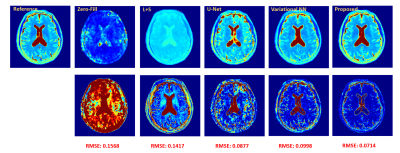
We demonstrated the proposed method in the setting of quantitative T2 mapping. Fully-sampled images were acquired from 85 subjects on a 3T scanner using the multi-echo spin-echo sequence with the following parameters: FOV=230×230×105 mm3, matrix size=256×256×21, TR=2000 ms, and TEs=[10.5,21,31.5,42,52.5,63] ms. The images were split into 75 and 10 subjects data for model training and testing, respectively. Sparse data were generated retrospectively using a random sampling mask with an acceleration factor of R=6.Figure 2 illustrates the effect of the priors used in the proposed method. As can be seen, both subspace and manifold priors largely reduced the aliasing artifacts over the zero-filled reconstruction, while their integration led to even greater improvement; the network-based sparsity prior picked up many residual features thus further improved the accuracy. Figure 3 compares the quality of the reconstructed images obtained by the existing methods (including both model-based and deep learning-based schemes) and the proposed method. Our method achieved the best reconstruction quality both qualitatively and quantitatively. The resulting T2 maps are shown in Fig. 4. Again, the proposed method obtained the most accurate results.
Conclusions
We present a new deep learning-based framework for synergistically integrating learned subspace, manifold, and sparsity priors to enable high-quality reconstruction of image sequences from highly sparse data. The proposed method has been validated using experimental data, producing impressive results. It may prove useful for many MRI applications that involve the acquisition of a sequence of images.Acknowledgements
This work reported in this paper was supported, in part, by NIH-R21-EB023413.References
[1] Liang Z-P. Spatiotemporal imaging with partially separable functions. In Proc IEEE Int Symp Biomed Imag. 2007;2:988–991.
[2] Lustig M, Santos JM, Donoho DL, Pauly JM. kt SPARSE: High frame rate dynamic MRI exploiting spatio-temporal sparsity. In Proc Intl Soc Magn Reson Med. 2006; 2420.
[3] Poddar S, Jacob M. Dynamic MRI using smoothness regularization on manifolds (SToRM). IEEE Trans Med Imag. 2015;35(4):1106-15.
[4] Lam F, Li Y, Peng X. Constrained magnetic resonance spectroscopic imaging by learning nonlinear low-dimensional models. IEEE Trans Med Imag. 2020;39(3): 545–555.
[5] Jung H, Sung K, Nayak KS, Kim EY, Ye JC. k-t FOCUSS: a general compressed sensing framework for high resolution dynamic MRI. Magn Reson Med. 2009;61(1):103–16.
[6] He J, Liu Q, Christodoulou AG, Ma C, Lam F, and Liang Z-P. Accelerated high-dimensional MR imaging with sparse sampling using low-rank tensors. IEEE Trans Med Imag. 2016;35(9):2119–2129.
[7] Li Y, Zhao Y, Guo R, Wang T, Zhang Y, Chrostek M, Low WC, Zhu XH, Liang Z-P, Chen W. Machine learning-enabled high-resolution dynamic deuterium MR spectroscopic imaging. IEEE Trans Med Imag. 2021.
[8] Lam F, Ma C, Clifford B, Johnson CL, Liang Z-P. High‐resolution 1H‐MRSI of the brain using SPICE: Data acquisition and image reconstruction. Magn Reson Med. 2016;76(4):1059-70.
[9] Chen Y, Li Y, Xu Z. Improved low-rank filtering of MR spectroscopic imaging data with pre-learnt subspace and spatial constraints. IEEE Trans Biomed Eng. 2019;67(8):2381-8.
[10] Aggarwal HK, Mani MP, Jacob M. MoDL: Model-based deep learning architecture for inverse problems. IEEE Trans Med Imaging. 2018;38(2):394-405.
[11] Hammernik K, Klatzer T, Kobler E, et al. Learning a variational network for reconstruction of accelerated MRI data. Magn Reson Med. 2018; 79(6): 3055-3071.
[12] Yang Y, Sun J, Li H, et al. ADMM-CSNet: A deep learning approach for image compressive sensing. IEEE Trans Pattern Anal Mach Intell. 2018; 42(3): 521-538.
[13] Schlemper J, Caballero J, Hajnal J V, et al. A deep cascade of convolutional neural networks for dynamic MR image reconstruction. IEEE Trans Med Imag. 2017; 37(2): 491-503.
Figures