3438
Automated Brain Extraction from T1-weighted MRI of Rhesus Macaques using a U-Net Deep Learning Framework1Biomedical Engineering, University of California, Davis, CA, United States, 2Radiology, University of California, Davis, CA, United States, 3Radiology, Wake Forest School of Medicine, Winston-Salem, NC, United States, 4California National Primate Research Center, University of California, Davis, CA, United States, 5Neurology, University of California, Davis, CA, United States
Synopsis
Transfer learning of an advanced deep learning framework, utilizing 3D U-Net pre-trained model, and trained on human brain MRI scans is proposed for brain extraction from MRI of other species. The proposed network architecture successfully performed whole brain extraction from rhesus macaque brain MRIs automatically with high accuracy, reduced errors, and lower computational cost. Options for data augmentation and different learning rates were also tested. Successful implementation of automated brain extraction would offer the potential to apply the same strategy on other animal models.
Introduction
Intensity-based brain extraction toolboxes have been used to perform automated human brain extraction for T1-weighted MRI. However, often they require manual edits because of inaccurate segmentation of the prefrontal lobes and inclusion of the skull when applying on non-human primates MRI scans. To overcome this problem, we propose a deep learning framework using the 3D U-Net1 and brain extraction weights computed from human MRI scans as the transfer learning model to perform accurate whole brain extraction without manual intervention.Method
Brain MRIs of thirteen rhesus macaque were acquired on a 3T Siemens Skyra (Siemens Healthineers, Erlangen, Germany) using a 4-channel monkey brain coil (Rapid Biomedical GmbH, Rimpar, Germany). A deep learning framework, Nobrainer2, was used for automated brain extraction. Datasets were converted to TFRecords to enable better compatibility with TensorFlow. T1-weighted MRIs (MP-RAGE: 3.65ms TE, 2.5s TR, 1.1s TI, 0.6mm isotropic resolution, 256x256 matrix size, 240 slices) were used as features and manually segmented brain masks were used as labels during training. Transfer learning was applied using a pre-trained 3D U-Net brain extraction model from 10,000 human brain MRIs. Twelve subjects were randomly chosen for transfer learning training and the remaining 1 subject was chosen for testing. For twelve training subjects, one pair of volumes are left for evaluation. Regularization and small transfer learning rate were applied to avoid dramatic changes in the learning parameter. Data augmentation using pure TensorFlow was tested at learning rate of the default rate, 1e-05. Then transfer learning at different learning rates (1e-03, 1e-04, 1e-05, 1e-06, 1e-07, and 1e-08) was also tested for further optimization with augmentation turned on. The brain from MRI of the test subject was extracted by using other intensity-based segmentation toolboxes (3dSkullStrip in AFNI and Brain Extraction Tool (BET) in FSL) for comparison. Dice coefficients of volumetric overlap between the reference and generated masks from the toolboxes and the proposed approach were calculated.Result
The processing time of the proposed method (20 minutes for training and 5 minutes for prediction) is significantly reduced compared to manual brain segmentation (2.5 hours). Our results showed that Nobrainer outperformed AFNI and FSL for the brain extraction for the rhesus brain MRIs. In particular, the brain mask that was created by transfer learning using Nobrainer showed better brain coverage, less inclusion of the non-brain tissues (Figure 1), and higher Dice coefficients than other toolboxes (Table 1). Brain masks obtained with data augmentation showed better results in terms of error reduction, brain coverage (Figure 2), and the highest Dice coefficient (Table 1). With data augmentation, transfer learning at learning rate of 1e-05 showed the best performance of generating the brain masks (Figure 3).Discussion
3D U-Net with transfer learning and data augmentation showed higher accuracy of brain extraction than common brain extraction tools such as AFNI and FSL. The proposed automated method is time-efficient without need of a species-specific template. The results showed that the transfer learning of brain extraction between human and non-human primates successfully estimated brain masks from rhesus brain MRI. This study may also have a potential for brain extraction in other species, with benefits to the preclinical brain imaging research community.Acknowledgements
NIH grants: RF1AG061001-01S1 (SSI/JHM) and R21AG064448 (AJC/JHM)
References
1. Ronneberger, Olaf, Philipp Fischer, and Thomas Brox. U-net: Convolutional networks for biomedical image segmentation. International Conference on Medical image computing and computer-assisted intervention. Springer, Cham 2015: 2344-2412.
2. Kaczmarzyk, Jakub, et al. Neuronets/Nobrainer: 0.1.0. Zenodo, 2021;0(1).
Figures
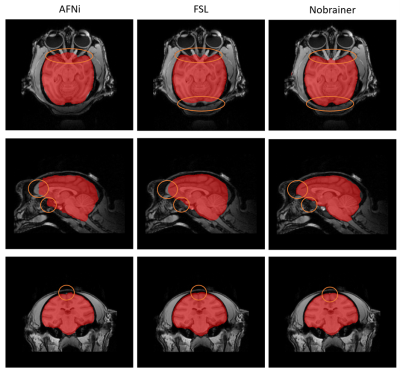
Figure 1. Comparison between brain masks obtained from AFNI (left), FSL (middle), and Nobrainer (right). Brain masks obtained from Nobrainer showed more complete brain coverage of prefrontal area and less error as shown by the orange circle.
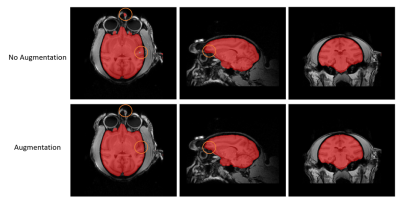
Figure 2. Comparison between brain masks obtained from Nobrainer transfer learning with data augmentation (top) and without data augmentation (bottom). Results showed that with data augmentation, brain masks obtained from Nobrainer transfer learning had less error as the orange circle indicated.
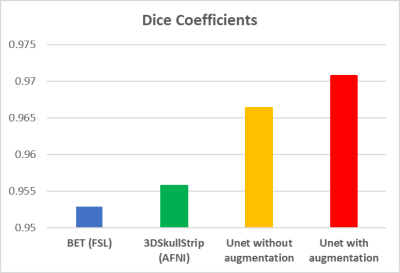
Table 1. Dice coefficients representing the volumetric overlap between the reference (manually segmented brain mask) and the brain masks generated using a toolbox.
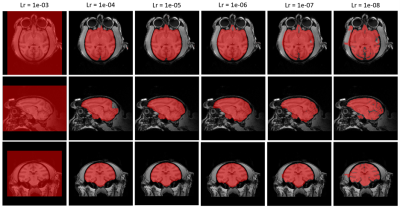
Figure 3. Comparison between brain masks obtained from Nobrainer transfer learning at different learning rate. Learning rate between 1e-05 and 1e-06 would display optimized completeness of brain extraction and minimized coverage of unwanted area.