3413
Rectal Cancer MRI Motion Quality Assessment using a Convolutional Neural Network1Radiology, Stanford University, Stanford, CA, United States, 2Electric Engineering, Stanford University, Stanford, CA, United States
Synopsis
Magnetic resonance (MR) imaging plays a pivotal role in the staging and treatment planning of rectal cancer. Accurate staging depends on good-quality high-resolution axial T2-weighted images orthogonal to the rectal tumor. Rectal MRI is often confounded by motion artifacts secondary to bowel peristalsis and patient movement. We propose a CNN model that automatically assesses image quality instantaneously after a scan is finished to reduce the frequency of patient recalls and non-diagnostic images. Our model achieves high accuracy in identifying motion degradation on an individual slice basis and perfect accuracy when classifying the entire sequence.
Introduction
Rectal cancer is one of the most common malignancies, with an incidence of 40 in 100,0001. High resolution orthogonal axial T2 weighted MRI imaging is essential for accurate staging of rectal cancer2,3. Specifically, this technique allows differentiation between rectal tumors confined within the rectal wall (stage T2 tumors) and those that extend beyond the muscularis propria (stage T3 tumors)4. Routine evaluation to assess for diagnostic quality imaging is often unable to be performed during the acquisition especially at high-volume imaging centers. We propose a neural network that provides immediate feedback to the technologist at the scanner on motion degradation of high-resolution axial T2 weighted imaging so that repeat imaging can be performed. This reduces the frequency of non-diagnostic imaging interpretation and patient callbacks thus improving patient care and satisfaction.Methods
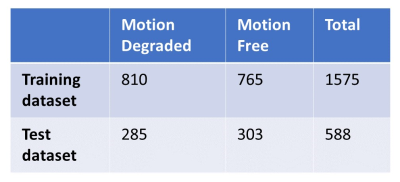
Rectal cancer MRIs performed on 3T MRI using a 32 channel phased array body coil at a single institution over a period of 13 months (October 2020 - October 2021) were analyzed retrospectively. The acquired dataset consisted of fully sampled 2D fast spin echo small field-of-view axial T2 weighted imaging prescribed perpendicular to the axis of the tumor with the following parameters: 3 mm slice thickness, TR 4000-6000, TE 100-150, FOV 16-20 cm, and matrix 256-320 x 256-320. 1 mg glucagon was administered intravenously immediately prior to the series acquisition. Within each collected series, the middle 60% was selected for model training as this was determined to routinely include the region of interest and exclude extraneous soft tissue and osseous structures not pertinent for evaluation of the rectal tumor for staging. The collected cases were divided into a training and a holdout test set. Our training set consisted of 63 cases containing 35 motion degraded and 28 motion free cases totaling 1575 slices. Our test set consisted of 24 cases containing 12 motion degraded and 12 motion free cases totaling 588 slices as shown in Table 1. Ground-truth labeling was separately performed by two radiologists applying binary pass/fail classification on each slice of the test set. Additionally, a single radiologist gave a binary pass/fail classification label on each slice in the training set. The binary pass/fail classification was defined as “pass” referring to a motion free image and “fail” referring to a motion degraded image. The utilized neural network was a multi-task model previously used to assess SNR and rigid motion corruption6. Data preprocessing and augmentation was performed. The model weights were initialized via transfer learning. Computations were performed using Python 3.7 with TensorFlow and Keras. A confusion matrix was constructed to compare the results using our model with those assigned by the radiologists.Results
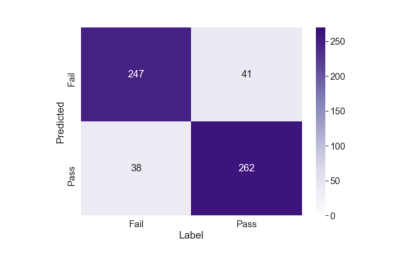
Inter-reader agreement between the two radiologists on the holdout test set was 89.6%. Figure 2 shows the confusion matrix obtained by the model (Predicted) and the ground truth (Label) by the radiologists on the test dataset. The proposed model demonstrated high performance with accuracy of 86.6% (509/588), precision of 87.3% (262/300), and recall of 86.5% (262/303). Additionally, the classification results of the individual slices were combined by using a majority vote aggregation function, meaning the most frequent slice within the sequence was assigned5. On a sequence-based analysis using the majority vote aggregation function, the model achieved an accuracy of 100%. The model correctly identified the 12 motion degraded cases warranting repeat imaging and the 12 motion free cases that were satisfactory for diagnostic evaluation.Discussion
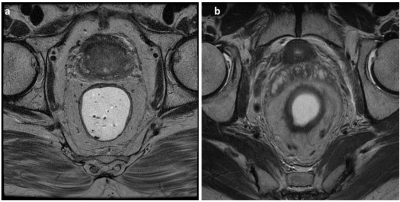
Rectal MRI is central to the staging and treatment planning of rectal cancer. Diagnostic accuracy is critically dependent on the image quality of a scan in order to adequately assess rectal tumors confined within the rectal wall versus those which extend beyond it4. The CNN architecture implemented in this study was able to classify images as motion degraded and motion free with a high degree of accuracy. An independent test set was used for the evaluation of model performance, so the high accuracy is unlikely related to overfitting. Combining the classification of individual slices with a majority vote aggregation function resulted in 100% accuracy for the classification of the entire series. This is likely secondary to the fact that artifacts affecting quality tend to be present on most slices of a sequence thus resulting in the high-performance accuracy. This circumstance translates well into the clinical environment when determining when repeating image series are warranted. Typical examples of motion free and motion degraded image slices are seen in Figure 1. We are planning to deploy the proposed model on clinical scanners and collecting feedback from the live environment. The application of our model can effectively improve clinical workflow by allowing the technologist to perform repeat imaging while the patient is still on the scanner therefore improving patient care and satisfaction and reducing costs.Conclusion
The study demonstrates a fully automated convolutional neural network achieving high accuracy in identifying motion degradation on an individual slice basis and perfect accuracy when classifying the entire high-resolution axial T2 weighted series. This model can help better optimize and ensure the acquisition of diagnostic rectal cancer MR imaging for accurate staging and treatment planning.Acknowledgements
No acknowledgement found.References
1. Maier A., Fuchsjager M.: Preoperative staging of rectal cancer. Eur J Radiol 2003; 47: pp. 89-97.
2. Taylor, Fiona GM, et al. "Preoperative magnetic resonance imaging assessment of circumferential resection margin predicts disease-free survival and local recurrence: 5-year follow-up results of the MERCURY study." Journal of Clinical Oncology 32.1 (2014): 34-43.
3. Furey, Elizabeth, and Kartik S. Jhaveri. "Magnetic resonance imaging in rectal cancer." Magnetic Resonance Imaging Clinics 22.2 (2014): 165-190.
4. Kaur, Harmeet, et al. "MR imaging for preoperative evaluation of primary rectal cancer: practical considerations." Radiographics 32.2 (2012): 389-409.
5. Cipollari, Stefano, et al. "Convolutional neural networks for automated classification of prostate multiparametric magnetic resonance imaging based on image quality." Journal of Magnetic Resonance Imaging (2021).
6. Lei K, Pauly J, Vasanawala S. Artifact- and content-specific quality assessment for MRI with image rulers. https://arxiv.org/abs/2111.03780, 2021.
Figures