3379
MRI lung lobe segmentation of pediatric cystic fibrosis patients using a neural network trained with publicly accessible CT datasets1Division of Radiological Physics, Department of Radiology, University Hospital Basel, University of Basel, Basel, Switzerland, 2Division of Pediatric Respiratory Medicine and Allergology, Department of Pediatrics, Inselspital, Bern University Hospital, University of Bern, Bern, Switzerland, 3Department of Biomedical Engineering, University of Basel, Basel, Switzerland, 4High Field Magnetic Resonance, Max Planck Institute for Biological Cybernetics, Tübinge, Germany, 5Department of Biomedical Magnetic Resonance, University of Tübingen, Tübingen, Germany, 6Department of Radiology, University Hospital Basel, University of Basel, Basel, Switzerland, 7Department of Diagnostic, Interventional and Pediatric Radiology, Inselspital, Bern University Hospital, University of Bern, Bern, Switzerland
Synopsis
Pulmonary biomarkers quantifications on a lobar level provide improved specificity against whole-lung analyses. However, lobar quantifications of pulmonary MR data are hardly accessible due to the complex work required for the manual segmentations. Supervised neural networks have shown the premise for automatic segmentation, but it is challenging to gather labelled data for the training. To overcome these limitations, in this work, we “translate” publicly accessible chest CT datasets and lobe segmentations to pseudo-MR data, and we then train a network able to segment consistently lung lobes of acquired MRI data. The cross-modality approach has excellent prospects to automatize MRI analyses.
Introduction and Purpose
Automated regional quantification of pulmonary biomarkers is of great interest for the assessment of respiratory diseases. For a superior regional analysis and an increased specificity, the quantification should be performed not only on the entire lung, but ideally also on a lobar level. In fact, the lungs are composed by five pulmonary lobes that can be considered independent functional units since they are invaginated in visceral pleurae, as well as supplied by different airways and vessels which do not extend from one lobe to another.Currently, lobar biomarker quantification of MR data is hardly accessible and rarely performed due to the cumbersome and complex work required for manual segmentation. Moreover, it is challenging to obtain a working algorithm or gather labelled data for supervised artificial neural network (ANN) training, and only a few studies exist.1–3
Supervised ANNs for segmentation or recognition tasks (e.g., lobes, vessels, pathologies) so far deliver superior results as compared to unsupervised or transfer learning ANNs, but are still generally hampered by the required large amounts of labelled datasets, often not available. However, labelled datasets from other imaging modalities or with different MR-contrasts might be available for supervised ANNs training, and can be directly employed once “translated” into the desired contrast.
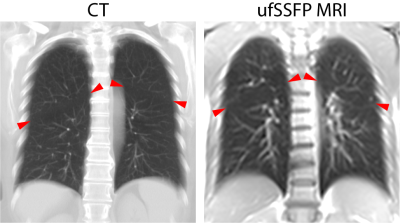
In this work, we propose a widely applicable workflow for MR lung lobe segmentation using a supervised recurrent neural network (RNN4,5) primarily trained with publicly available CT data and lobe masks, rather than MR images and manually labelled MR lobe masks. The feasibility is demonstrated for 2D coronal ultra-fast balanced steady-state free precession (ufSSFP) MRI, generally used for free-breathing and contrast agent-free ventilation and perfusion imaging with matrix pencil decomposition.6,7
Methods
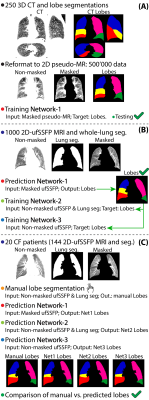
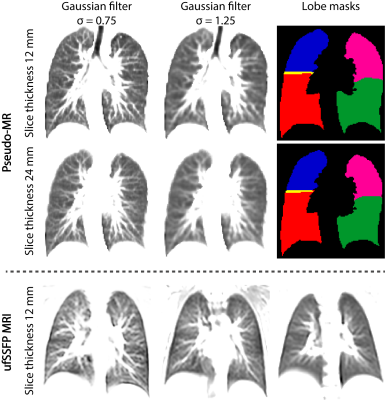
A schematic of the study workflow is presented in Figure 1. Publicly accessible chest CT data8 of 250 adult patients with suspected pulmonary nodules were used and the lung lobes were segmented with the open-source CNN of Hofmanninger et al.9 To match ufSSFP data of pediatric patients, both CT data and segmentations were translated into pseudo-MR images with specific image processing steps. Further, the images were masked to suppress the anatomy outside the lung regions since exhibiting strongly different image features and signal intensities from MR (Figure 2). Exemplary pseudo-MR images are shown in Figure 3, and more details are in preprint on arXiv.10We trained and tested 3 Networks (Figure 1). Network-1 was trained with pseudo-MR images and lobe segmentations, and applied to 1000 masked ufSSFP images to predict lobe segmentations. These outputs were directly used as targets to train Network-2 and Network-3 with non-masked ufSSFP data as inputs, and an additional whole-lung mask as input for Network-2. Whole-lung segmentations of MR data were performed with a previously trained RNN.11,12
Network predictions were compared to reference manual lobe segmentations of ufSSFP data in twenty pediatric cystic fibrosis patients.
Results
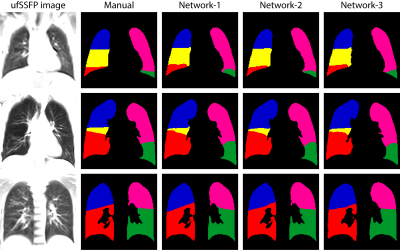
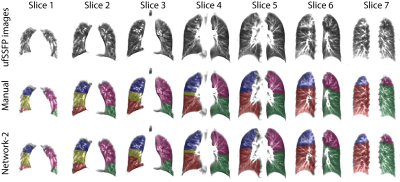
Network-1 was able to segment the lobes of ufSSFP images, and Network-2 and Network-3 further increased segmentation accuracy and robustness. Representative segmentations are shown in Figure 4. The average all-lobe Dice similarity coefficients were 95.0±2.3 (mean±pooled SD [%]), 96.4±1.2, 93.0±1.8, and the average median Hausdorff distances were 6.1±0.9 (mean±SD [mm]), 5.3±1.1, 7.1±1.3, for Network-1, Network-2, and Network-3, respectively. The lung lobe volumes for networks and manual segmentations were very highly correlated (r>0.90, p<10-7).A whole-lung ufSSFP examination of a CF subject along with manual and Network-2 predicted segmentations is illustrated in Figure 5. The inferred lobe segmentations with Network-2 have a good similarity with the manual masks and an excellent inter-slice lobe-mask consistency.
Discussion
We introduced a widely applicable workflow that provides effective lobe segmentation of MR images without using either ground-truth MR labelled datasets or a multitude of MR data (Network-1). We further improved the RNN performance and robustness with Network-2 and Network-3, without performing either data selection or mask refinements to develop a novel workflow without need for user interaction.Our workflow requires an available whole-lung segmentation, but this is in fact a prerequisite for all pulmonary quantification processes. Moreover, nowadays, segmentation of relatively simple organs such as the whole lung might be feasible with only a limited number of datasets by using specific ANNs.13 At the cost of increased complexity, deep learning‐based imaging synthesis could be adopted to translate chest CT images into pseudo-MR images,14 possibly eliminating the need for whole-lung segmentations.
The proposed cross-modality, cross-pathology, and cross-age approach might be relevant to foster further MRI pulmonary research, especially in children, in which CT is preferably avoided. We expect that the proposed workflow is broadly applicable and can easily be adapted to quantify pulmonary biomarkers on several MRI acquisitions (e.g., GRE, UTE, DCE-MRI, 2D/3D). Moreover, we hypothesize that our method might be adaptable to segment the lung vessels or other pathologies (e.g., nodules, embolisms) on MR data, starting from CT labelled datasets. In general, CT-to-MR image translation has excellent prospects to automatize pulmonary MR data analysis and to accelerate the clinical inclusion of lung MRI, potentially improving respiratory health care.
Conclusion
RNN lung lobe segmentation of 2D ufSSFP imaging is feasible, in good agreement with manual segmentations. The proposed workflow might provide rapid access to automated lobe segmentation for various lung MRI examinations and quantitative analyses.Acknowledgements
Orso Pusterla acknowledges the support of the Swiss Cystic Fibrosis Society (CFCH).References
1. Tustison NJ, Qing K, Wang C, Altes TA, Mugler JP. Atlas-based estimation of lung and lobar anatomy in proton MRI. Magnetic Resonance in Medicine. 2016:315-320.
2. Lee H, Matin T, Gleeson F, Grau V. Learning-Based Multi-atlas Segmentation of the Lungs and Lobes in Proton MR Images. In: Descoteaux M, Maier-Hein L, Franz A, Jannin P, Collins DL, Duchesne S, eds. Medical Image Computing and Computer Assisted Intervention − MICCAI 2017. Springer International Publishing; 2017:108-115.
3. Winther HB, Gutberlet M, Hundt C, et al. Deep semantic lung segmentation for tracking potential pulmonary perfusion biomarkers in chronic obstructive pulmonary disease (COPD): The multi-ethnic study of atherosclerosis COPD study. J Magn Reson Imaging. 2020;51(2):571-579. doi:10.1002/jmri.26853.
4. Andermatt S, Pezold S, Cattin P. Multi-dimensional Gated Recurrent Units for the Segmentation of Biomedical 3D-Data BT - Deep Learning and Data Labeling for Medical Applications: First International Workshop, LABELS 2016, and Second International Workshop, DLMIA 2016, Held in Conjunctio. In: Carneiro G, Mateus D, Peter L, et al., eds. Springer International Publishing; 2016:142-151. doi:10.1007/978-3-319-46976-8_15.
5. Andermatt S, Pezold S, Cattin PC. Automated segmentation of multiple sclerosis lesions using multi-dimensional gated recurrent units. In: Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics). Vol 10670 LNCS. Springer, 2017; 2018:31-42. doi:10.1007/978-3-319-75238-9_3.
6. Bauman G, Pusterla O, Bieri O. Ultra-fast Steady-State Free Precession Pulse Sequence for Fourier Decomposition Pulmonary MRI. Magnetic Resonance in Medicine. 2015:75:1647-53.
7. Bauman G, Bieri O. Matrix pencil decomposition of time-resolved proton MRI for robust and improved assessment of pulmonary ventilation and perfusion. Magn Reson Med. 2017;77(1):336-342. doi:10.1002/mrm.26096.
8. Setio AAA, Traverso A, de Bel T, et al. Validation, comparison, and combination of algorithms for automatic detection of pulmonary nodules in computed tomography images: The LUNA16 challenge. Med Image Anal. 2017;42:1-13. doi:https://doi.org/10.1016/j.media.2017.06.015.
9. Hofmanninger J, Prayer F, Pan J, Röhrich S, Prosch H, Langs G. Automatic lung segmentation in routine imaging is primarily a data diversity problem, not a methodology problem. Eur Radiol Exp. 2020;4(1):50. doi:10.1186/s41747-020-00173-2.
10. Pusterla O, Heule R, Santini F, et al. MRI lung lobe segmentation in pediatric cystic fibrosis patients using a recurrent neural network trained with publicly accessible CT datasets. ArXiv. Published online 2021. https://doi.org/10.1007/s10334-019-00754-2.
11. Pusterla O, Andermatt S, Bauman G, et al. Deep Learning Lung Segmentation in Paediatric Patients. In: Proc. Intl. Soc. Mag. Reson. Med. 26; 2018:4355.
12. Willers C, Bauman G, Andermatt S, et al. The impact of segmentation on whole-lung functional MRI quantification: Repeatability and reproducibility from multiple human observers and an artificial neural network. Magn Reson Med. Published online September 2020. doi:10.1002/mrm.28476.
13. Cheplygina V, de Bruijne M, Pluim JPW. Not-so-supervised: A survey of semi-supervised, multi-instance, and transfer learning in medical image analysis. Med Image Anal. 2019;54:280-296. doi:10.1016/j.media.2019.03.009.
14. Wang T, Lei Y, Fu Y, et al. A review on medical imaging synthesis using deep learning and its clinical applications. J Appl Clin Med Phys. 2021;22(1):11-36. doi:https://doi.org/10.1002/acm2.13121.
Figures