3369
Probing Liver Microstructure in-vivo Using Diffusion-Relaxation Correlation Spectroscopic Imaging (DR-CSI)1Department of Radiology, University of Southern California, Los Angeles, CA, United States, 2Department of Medicine, Division of Gastroenterology and Liver Diseases, University of Southern California, Los Angeles, CA, United States, 3Department of Bioengineering, University of California, Los Angeles, Los Angeles, CA, United States, 4Department of Electrical and Computer Engineering, University of Southern California, Los Angeles, CA, United States, 5Department of Biomedical Engineering, University of Southern California, Los Angeles, CA, United States, 6Department of Radiation Oncology, University of Southern California, Los Angeles, CA, United States
Synopsis
Diffusion-relaxation correlation spectroscopic imaging (DR-CSI) is an advanced microstructure imaging approach that can resolve sub-voxel tissue compartments and quantify their fractions. In this work, we investigated the feasibility of DR-CSI with an optimized experiment design for in-vivo liver imaging. Our study showed that DR-CSI can measure multiple sub-voxel compartments in the liver and provide consistent component fraction maps in healthy livers. An initial test on a subject with chronic hepatitis B also demonstrated the potential of DR-CSI to identify and characterize pathological changes in liver parenchyma. Further studies on variable liver diseases are underway.
Introduction
Quantitative multiparametric MR imaging has been of great interest to provide important biomarker to understand various liver pathologies1. While multiparametric mapping approaches take advantage of rich information from multiple MR contrast mechanisms, they are limited in probing multiple sub-voxel compartments of the liver because of the single-compartment assumption. Recently, high-dimensional spectrum approaches2,3 have been shown to be capable of resolving sub-voxel compartments in various human imaging applications, for example, in the brain4,5, the prostate6, the placenta7, etc. Diffusion-relaxation correlation spectroscopic imaging (DR-CSI) is one such approach that acquires multidimensional data with the simultaneous encoding of diffusion and relaxation and estimates a 2D diffusion-relaxation correlation spectrum for every voxel in which multiple peaks corresponding to different sub-voxel compartments are observed2,4. This method has not been investigated in the liver, which could be potentially useful for better understanding of liver microstructure. In this work, we demonstrate the feasibility of DR-CSI with an optimized experiment design for in-vivo liver imaging.Methods
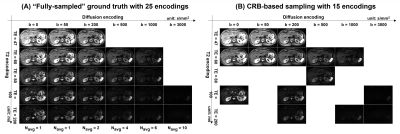
Data acquisition: Seven healthy livers and one liver infected with the hepatitis B virus were scanned with IRB approval. For each subject, abdominal DR-CSI data were acquired using a free-breathing diffusion-weighted spin-echo EPI sequence at 3T (MAGNETOM Vida, Siemens) with TR=7000ms, voxel size=1.6x1.6x5mm3, 35 slices, and 3-scan trace weighted diffusion encoding for each b-value. As shown in Figure 1, a total of 25 combinations from 6 b-values (b = 0,50,200,500,1000 and 3000 s/mm2) and 5 echo times (TE = 47,60,80,100 and 120ms) were used for contrast encoding as a “fully-sampled” ground truth (total scan time = 28 min). From the 25 contrast encodings, a total of 15 combinations were selected using the Cramer-Rao Bound (CRB)-based experiment design8-9 to shorten the scan time to 18min.Data analysis: DR-CSI assumes a multi-compartment signal model defined by:
$$M(x,y,b,TE) = \int\int F(x,y,D,T_2) e^{-bD}e^{-TE/T_2}~dD~dT_2,$$
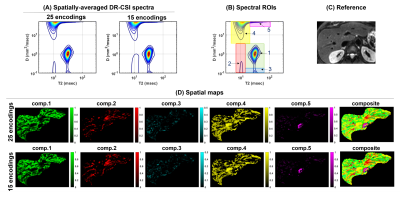
where $$$M(x,y,b,TE)$$$ is the measured diffusion-weighted spin-echo data and $$$F(x,y,D,T_2) $$$ is the 2D diffusion-T2 correlation spectrum at location $$$(x,y)$$$. The DR-CSI spectra from all voxels are estimated by solving a dictionary-based spatially-regularized nonnegative least squared optimization problem 2,3. For the analysis, we defined spectral ROIs of a rectangular shape (Fig. 2B) for distinct spectral peaks that represent different sub-voxel compartments. Spatial maps were generated for each spectral ROIs that indicate the fractions of individual sub-voxel compartments at each voxel.
Results
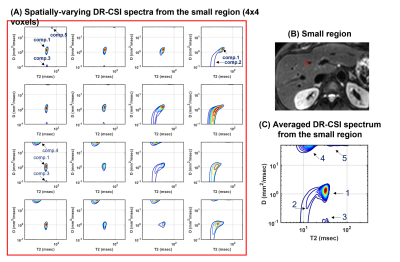
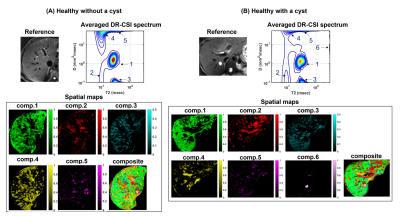
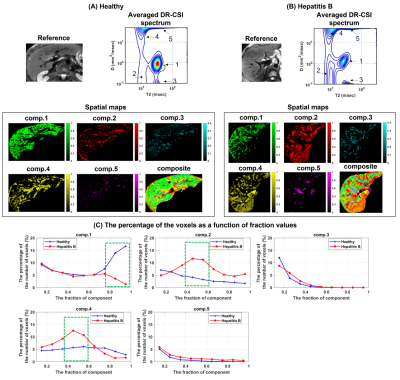
Figures 2 and 3 show the results from one healthy subject. The ground truth (25 encodings) and the accelerated acquisition (15 encodings) are compared in Fig. 2(A) and 2(D). We observed consistent five spectral peaks in the spatially-averaged DR-CSI spectra in (A). The spectral ROIs for these peaks are defined in (B) and the fraction maps corresponding to these ROIs are shown in (D). The fraction maps of the accelerated acquisitions were in accordance with the maps from the ground truth. These components seem to correspond to hepatocytes (comp.1), bile ducts (comp.2), connective tissues (comp.3) and a part of blood vessels (comp.4 and comp.5). Fig. 3 shows spatially-varying DR-CSI spectra in a small region from the accelerated scan, clearly showing the transitions of the five components and the partial-volume effects of them. Figure 4 shows the DR-CSI results from another two healthy livers with the accelerated scan (15 contrast encodings), showing good consistency across different subjects. Furthermore, Fig. 4(B) shows one additional spectral peak (component 6) that is not present in other subjects. This component seems to correspond to a cyst, which demonstrates an ability of DR-CSI to detect abnormality. Figure 5 shows the comparisons between a healthy liver and a liver infected by the hepatitis B virus. Five components were observed in both livers. However, the fractions of the components, in particular components 1, 2 and 4, are dramatically different. This suggests that DR-CSI has the potential to identify and characterize pathological changes in liver parenchymaConclusion
We demonstrated that DR-CSI can be feasible for in-vivo liver microstructure imaging with a clinically practical data acquisition time. Our results suggest that DR-CSI has abilities of resolving liver sub-voxel compartments, capturing abnormality, and/or potentially characterizing clinically important pathological changes. Validating the liver sub-voxel compartments and evaluating its utility with various liver diseases are our ongoing work.Acknowledgements
No acknowledgement found.References
1. Banerjee R, Pavlides M, Tunnicliffe EM, Piechnik SK, Sarania N, Philips R, Collier JD, Booth JC, Schneider JE, Wang LM, Delaney DW, Multiparametric magnetic resonance for the non-invasive diagnosis of liver disease. J Hepatol, 2014:60;69-77.
2. Kim D, Doyle EK, Wisnowski JL, Kim JH, Haldar JP, Diffusion‐relaxation correlation spectroscopic imaging: a multidimensional approach for probing microstructure. Magn Reson Med. 2017:78;2236-2249.
3. Slator PJ, Palombo M, Miller KL, Westin CF, Laun F, Kim D, Haldar JP, Benjamini D, Lemberskiy G, de Almeida Martins JP, Hutter J. Combined diffusion‐relaxometry microstructure imaging: Current status and future prospects. Magn Reson in Med. 2021 (online).
4. Kim D, Wisnowski JL, Nguyen CT, Haldar JP, Multidimensional correlation spectroscopic imaging of exponential decays: from theoretical principles to in vivo human applications. NMR in Biomed. 2020:33;e4244.
5. Benjamini D, Iacono D, Komlosh ME, Perl DP, Brody DL, Basser PJ. Diffuse axonal injury has a characteristic multidimensional MRI signature in the human brain. Brain. 2021;144:800-16.
6. Zhang Z, Wu HH, Priester A, Magyar C, Afshari Mirak S, Shakeri S, Mohammadian Bajgiran A, Hosseiny M, Azadikhah A, Sung K, Reiter RE, Prostate microstructure in prostate cancer using 3-T MRI with diffusion-relaxation correlation spectrum imaging: validation with whole-mount digital histopathology. Radiology. 2020:296;348-355.
7. Slator PJ, Hutter J, Palombo M, Jackson LH, Ho A, Panagiotaki E, Chappell LC, Rutherford MA, Hajnal JV, Alexander DC. Combined diffusion‐relaxometry MRI to identify dysfunction in the human placenta. Magn Reson in Med. 2019;82:95-106.
8. Kim D, Haldar JP. Faster Diffusion-Relaxation Correlation Spectroscopic Imaging (DR-CSI) using Optimized Experiment Design, ISMRM. 2017;176.
9. Kim D, Wisnowski JL, Haldar JP. Improved Efficiency for Microstructure Imaging using High-Dimensional MR Correlation Spectroscopic Imaging. In Proceedings of Asilomar Conference on Signals, Systems and Computers. 2017; 1264-1268.
Figures