3284
Applying the single shell 3-Tissue method to investigate long-term degeneration of the visual system following hemidisconnection surgery1Developmental Imaging and Biophysics Section, UCL Great Ormond Street Institute of Child Health, LONDON, United Kingdom, 2Children’s Hospital of Pittsburgh, University of Pittsburgh Medical Centre, Pittsburgh, KS, United States, 3Clinical and Academic Department of Ophthalmology, Great Ormond Street Hospital for Children NHS Foundation Trust, London, United Kingdom, 4Neurosurgery, Great Ormond Street Hospital for Children NHS Foundation Trust, London, United Kingdom, 5Clinical Neurosciences, UCL Great Ormond Street Institute of Child Health, LONDON, United Kingdom
Synopsis
There is a need to map microstructural changes over long time periods and develop/apply methods that work with legacy data. In this study, we applied the novel single shell 3-Tissue method to data from a cohort of 4 patients who were scanned 20-years following childhood hemidisconnection surgery and presented degeneration of the visual system. We believe this study suggests that diffusion MRI can be used to monitor the integrity of the visual system following hemispherectomy and if extended to larger cohorts and a greater number of time-points, provide a clearer picture of the natural history of visual system degeneration.
Introduction
Diffusion tensor imaging (DTI) (1) and tractography are tools currently used in many clinical settings, including neurosurgical planning and evaluation of microstructural integrity following surgery (2,3). Nonetheless, DTI has inherent limitations in capturing complex microstructural arrangements where diffusion of water molecules does not follow a gaussian distribution. Recent advances in scanning and modelling technologies have enabled to move beyond the limitations of DTI, but the challenge still remains to apply some of those techniques to legacy data. Therefore, in this study we have applied the single shell 3-Tissue method (4) to investigate long-term degeneration of the visual system following hemidisconnection surgery.Methods
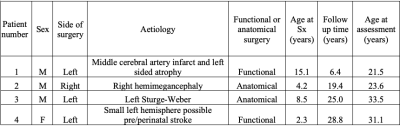
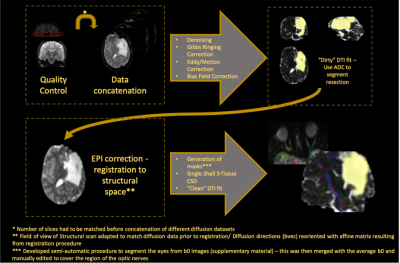
Legacy data for 4 patients (clinical details available in Figure 1) was available including: Echo-planar diffusion weighted images with 20 non-collinear directions, b = 1000 s.mm−2 and single b0 volume, 45 contiguous axial slices of thickness 2.5 mm per dataset, FOV= 240×240 mm and 96×96 voxel acquisition matrix, for a final image resolution of 2.5×2.5×2.5 mm, TE=89 ms, TR=6300 ms, NEX=3. In addition, an identical protocol with 60 non-collinear gradient directions and three b0 volumes was acquired. In order to increase SNR and angular coverage, both sets of diffusion scans were concatenated following visual assessment and discarding of corrupted volumes. A T1-weighted 3D FLASH structural image was also available with 176 contiguous sagittal slices, FOV=256×224 mm, Flip Angle=15°, TE=4.9 ms, TR=11 ms and 1×1×1 mm image resolution.Pre-processing included denoising (5), Gibbs ringing correction and eddy current and motion distortion correction using MRtrix3 (6) and FSL tools (7). EPI distortion correction was then performed by registering the average b0 scan to the structural scan with the Advanced Normalisation Tools Software (ANTs) (8). After EPI correction we applied the single shell 3-Tissue constrained spherical deconvolution to extract fibre orientation distribution functions for our data using the MRtrix3Tissue tool (https://3Tissue.github.io). Finally, tractography of the complete (intact) visual pathways (including both optic nerves, contra lateral optic tract and contra lateral optic radiation) was performed for all patients, with the following details: CSD probabilistic tractography (iFOD2): angular threshold = 90°, 5000 streamlines; FA threshold = 0.15; FOD amplitude = 0.1.
After tractography maps were obtained, each individual diffusion dataset that was concatenated before pre-processing was split back according to the original distribution (as described above) and the DTI model applied so that metrics could be extracted from every white matter structure (complete image analysis pipeline shown in figure 2). Purely for qualitative comparison of the obtained results, we have additionally reconstructed metrics of interest in a group of 20 age-matched healthy controls.
Results
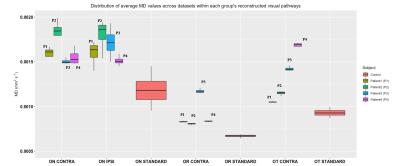
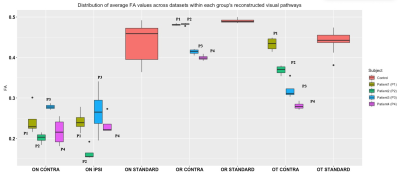
Qualitatively we can appreciate that for all patients and all components of the visual system (Optic nerves, contra lateral optic tract and contra lateral optic radiation) there are abnormalities characterized by increases in mean diffusivity and reductions in fractional anisotropy (Figure 3 and 4). In all patients, both MD and FA are very similar between the ipsi- and contralateral optic nerves. However, analysis of the remaining structures in the contralateral hemisphere showed differences in FA and MD across patients. Finally, reconstructed values within the optic radiation and optic tract were more reproducible across the available diffusion datasets whereas the optic nerves showed higher variability.Discussion
Despite the low variability found in the reconstruction of DTI metrics within the optic nerves, it is well known that these structures are very challenging to image and therefore further work on obtaining high-resolution imaging of the optic nerves would be valuable to evaluate any structural differences between the optic nerves that could explain the inter ocular differences found in the ophthalmological parameters (data not shown). Furthermore, although we did not employ a formal statistical comparison with the healthy control group, we recognize that there can be a systematic bias in FA, which is small when compared to the magnitude of the changes that we observe. Future work looking at harmonisation techniques (9) would provide insights in how to overcome these biases.Conclusion
Our study has shown the usefulness of the single shell 3-Tissue method to reconstruct white matter pathways from legacy data beyond the limitations of DTI-based tractography. Working with legacy datasets is unavoidable but with techniques such as the one used here we have shown that we may be able to gather a large body of evidence to more robustly explore the degeneration of the visual system following hemidisconnection surgery. This can have very important implications in advising patients before a surgical decision is made, and also to be used as outcome measure of the impact of rehabilitation strategies.Acknowledgements
No acknowledgement found.References
1 - Basser, P. J., Mattiello, J., & LeBihan, D. (1994). MR diffusion tensor spectroscopy and imaging. Biophysical Journal, 66(1), 259–267. https://doi.org/10.1016/S0006-3495(94)80775-1
2 - Winston, G. P., Daga, P., White, M. J., Micallef, C., Miserocchi, A., Mancini, L., Modat, M., Stretton, J., Sidhu, M. K., Symms, M. R., Lythgoe, D. J., Thornton, J., Yousry, T. A., Ourselin, S., Duncan, J. S., & McEvoy, A. W. (2014). Preventing visual field deficits from neurosurgery. Neurology, 83(7), 604–611. https://doi.org/10.1212/WNL.0000000000000685
3 - Lacerda, L. M., Clayden, J. D., Handley, S. E., Winston, G. P., Kaden, E., Tisdall, M., Cross, J. H., Liasis, A., & Clark, C. A. (2020). Microstructural Investigations of the Visual Pathways in Pediatric Epilepsy Neurosurgery: Insights From Multi-Shell Diffusion Magnetic Resonance Imaging. Frontiers in Neuroscience, 14. https://doi.org/10.3389/fnins.2020.00269
4 - Dhollander, T., & Connelly, A. (2016). A novel iterative approach to reap the benefits of multi-tissue CSD from just single-shell (+b=0) diffusion MRI data. International Society for Magnetic Resonance in Medicine.
5 - Veraart, J., Novikov, D. S., Christiaens, D., Ades-aron, B., Sijbers, J., & Fieremans, E. (2016). Denoising of diffusion MRI using random matrix theory. NeuroImage, 142, 394–406. https://doi.org/10.1016/j.neuroimage.2016.08.016
6 - Tournier, J.-D., Smith, R., Raffelt, D., Tabbara, R., Dhollander, T., Pietsch, M., Christiaens, D., Jeurissen, B., Yeh, C.-H., & Connelly, A. (2019). MRtrix3: A fast, flexible and open software framework for medical image processing and visualisation. NeuroImage, 202, 116137. https://doi.org/10.1016/j.neuroimage.2019.116137
7 - Jenkinson, M., Beckmann, C. F., Behrens, T. E. J., Woolrich, M. W., & Smith, S. M. (2012). FSL. NeuroImage, 62(2), 782–790. https://doi.org/10.1016/j.neuroimage.2011.09.015
8 - Avants, B. B., Tustison, N. J., Stauffer, M., Song, G., Wu, B., & Gee, J. C. (2014). The Insight ToolKit image registration framework. Frontiers in Neuroinformatics, 8. https://doi.org/10.3389/fninf.2014.00044
9 - Pinto, M. S., Paolella, R., Billiet, T., Van Dyck, P., Guns, P.-J., Jeurissen, B., … Sijbers, J. (2020). Harmonization of Brain Diffusion MRI: Concepts and Methods. Frontiers in Neuroscience, 14. https://doi.org/10.3389/fnins.2020.00396
Figures