3255
Machine learning to detect microstructural brain changes in patients with amnestic mild cognitive impairment based on NODDI1Department of Radiology, Tianjin Medical University General Hospital, Tianjin, China, 2Department of Radiology, First Central Clinical institution, Tianjin Medical University, Tianjin, China, 3Institute of Medical Engineering and Translational Medicine, Tianjin University, Tianjin, China, 4MR Collaboration, Siemens Healthineers Ltd., Beijing, China, 5Department of Radiology, Tianjin First Central Hospital, Tianjin, China
Synopsis
This study investigated the changes in brain microstructure in patients with amnestic mild cognitive impairment (aMCI) using neurite orientation dispersion and density imaging (NODDI) combined with machine learning. Neurite density index (NDI) was significantly decreased in white matter, orientation dispersion index (ODI) was significantly decreased in gray matter, and volume fraction of isotropic water molecules (Viso) was significantly increased in the aMCI group. Further correlation and receiver operating characteristic (ROC) curve analyses showed NODDI may reflect the clinical cognitive status of aMCI. NODDI combined with a machine learning algorithm could be a promising alternative for early diagnosis of MCI.
Introduction
At present, the treatment window of Alzheimer’s Disease (AD) has moved forward to mild cognitive impairment (MCI) and even the pre-dementia stage. Early diagnosis of MCI is crucial in slowing down the progression of dementia. Many studies based on MRI diffusion tensor imaging (DTI) have shown that the fractional anisotropy (FA) values decrease in MCI as the myelin sheath is damaged 1. However, FA is nonspecific as it is also related to axon diameter and density, fiber distribution, and partial volume effect. Neurite orientation dispersion and density imaging (NODDI) is a multi-compartment diffusion model based on the difference of water molecule diffusion in intracellular, extracellular, and cerebrospinal fluid 2. Compared with conventional DTI, the brain microstructure can be more accurately assessed with NODDI. The rapidly development of machine learning algorithms has been widely studied in diagnostic assistance and disease prognosis 3. Therefore, the purpose of this study was to investigate the changes in brain microstructure in patients with amnestic MCI (aMCI) by using NODDI combined with machine learning.Methods
1. Data AcquisitionThis study consisted of 26 aMCI patients and 24 age, gender, and education-year matched normal controls. aMCI patients fulfilled the Petersen criteria and Mini-Mental State examination (MMSE) score≥24, Montreal Cognitive Assessment (MoCA) score<26, and Clinical dementia rate (CDR)=0.5. Following informed consent, all subjects were scanned on a 3T MAGNETOM Trio a Tim System (Siemens Healthcare, Erlangen, Germany) with a 32-channel head coil. Whole-brain diffusion-weighted images for NODDI analysis were collected using a prototype multi-band echo planar imaging sequence with the following parameters: TR/TE=4600/95 ms, FOV=220×220 mm2, voxel size=2×2×2 mm3, 70slices, multi-band acceleration factor=2, diffusion directions=64, b = 0/1000/2000 s/mm2.
2. Data Analysis
Head motion and eddy currents were corrected in FSL (https://fsl.fmrib.ox.ac.uk/fsl/fslwiki/). The corrected data was processed by NODDI_toolbox (https://www.nitrc.org/projects/noddi_toolbox/) and the NODDI parameters including neurite density index (NDI), orientation dispersion index (ODI) and volume fraction of isotropic water molecules (Viso), were obtained. Based on the white matter (WM) atlas of Johns Hopkins University, 11 templates of white matter structures were made, including the bilateral cingulum, genu, and splenium of corpus callosum, bilateral posterior limb of internal capsule, bilateral superior longitudinal fasciculus, bilateral uncinate fasciculus and fornix. 14 templates of gray matter (GM) structures were made based on anatomical automatic labeling template (AAL), which included bilateral hippocampus, parahippocampal gyrus, amygdala, caudate nucleus, globus pallidus, putamen and thalamus. The NDI, ODI, and Viso values of all the templates were extracted. Multiple machine learning algorithms, including K-nearest neighbor (KNN), logistic regression (LR), random forest (RF) and support vector machine (SVM), were tested to evaluate the diagnostic efficiency of each parameter value on aMCI. The extracted templates values were tested between the two groups by independent sample t-test and the correlation between the structures with statistical difference and Mini-Mental State examination (MMSE) and Montreal Cognitive Assessment (MoCA) scores were analyzed.
Results
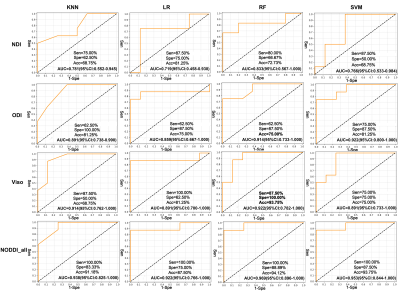
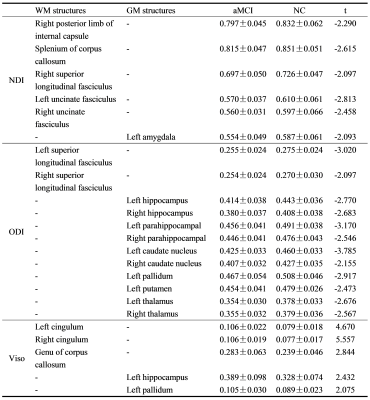
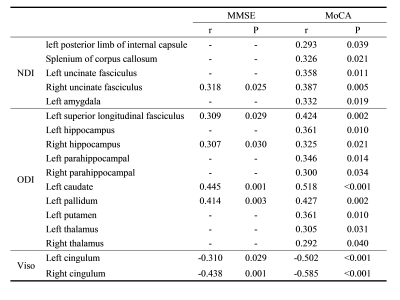
Using the KNN, LR, RF, and SVM machine learning algorithms, the areas under the ROC curve (AUC) of predicting aMCI by the NDI values of all the templates were 0.781, 0.719, 0.833, and 0.766 respectively; the AUCs of predicting aMCI by the ODI values of all the templates were 0.891, 0.859, 0.914, and 0.922 respectively; the AUCs of predicting aMCI by the Viso values of all the templates were 0.914, 0.891, 0.922, and 0.891 respectively; the AUCs of predicting aMCI by all the NODDI parameters of all the templates (NODDI_all) were 0.938, 0.922, 0.969, and 0.953 respectively (Figure 1). In WM, the NDI values of 45% (5/11) and the ODI values of 18% (2/11) decreased, and the Viso values of 27% (3/11) increased in the aMCI group. While in the GM, the NDI values of 7% (1/14) and the ODI values of 71% (10/14) decreased, and the Viso values of 14% (2/14) increased in the aMCI group (Table 1). The NDI value of right uncinate fasciculus, the ODI values of left superior longitudinal fasciculus, right hippocampus, left caudate nucleus, and left pallidum, and the Viso values of bilateral cingulum and left hippocampus significantly correlated with MMSE score (Table 2). The NDI values of the splenium of corpus callosum, bilateral uncinate fasciculus and left amygdala, the ODI values of the left superior longitudinal fasciculus, bilateral hippocampus, left parahippocampal gyrus, left caudate nucleus, left pallidum, left putamen and left thalamus, the Viso values of bilateral cingulum and left hippocampus significantly correlated with MoCA scores (Table 2).Discussion and Conclusion
This study evaluated the ability of NODDI to detect brain microstructure changes of aMCI. Compared with normal controls, NDI and ODI were significantly decreased and Viso was significantly increased in the aMCI group. The results indicated that the decrease of neurite density was the main change in WM and the decrease of dendritic complexity was the main change in GM in aMCI patients. The correlation analysis suggests that NODDI may reflect the clinical cognitive status of patients with aMCI. NODDI combined with machine learning algorithm was expected to be a novel method for early diagnosis of MCI.Acknowledgements
No acknowledgement foundReferences
1. Yu J, Lam C L M, Lee T M C. White matter microstructural abnormalities in amnestic mild cognitive impairment: A meta-analysis of whole-brain and ROI-based studies[J]. Neurosci Biobehav Rev, 2017,83:405-416.
2. Zhang H, Schneider T, Wheeler-Kingshott C A, et al. NODDI: practical in vivo neurite orientation dispersion and density imaging of the human brain[J]. Neuroimage, 2012,61(4):1000-1016.
3. Jo T, Nho K, Saykin A J. Deep Learning in Alzheimer's Disease: Diagnostic Classification and Prognostic Prediction Using Neuroimaging Data[J]. Front Aging Neurosci, 2019,11:220
Figures