3215
Ultrafast water-fat separation using deep learning-based single-shot MRI1Department of Electronic Science, Xiamen University, Xiamen, China, 2Department of Biomedical Engineering, City University of Hong Kong, Hong Kong, China
Synopsis
Water-fat separation is a powerful tool in diagnosing many diseases and many efforts have been made to reduce the scan time. Spatiotemporally encoded (SPEN) single-shot MRI, as an emerging ultrafast MRI method, can accomplish the fastest water-fat separation since only one shot is required. However, the SPEN water/fat images obtained by the state-of-the art methods still have some shortcomings. Here, a deep learning approach based on U-Net was proposed to obtain SPEN water/fat images simultaneously with improved spatial resolution, better fidelity and reduced reconstruction time. The efficiency of our method is demonstrated by numerical simulations, and in vivo rat experiments.
Introduction
Water-fat separation is a powerful tool in diagnosing many diseases and has been increasingly used in clinical practices.1 Currently, most of the water-fat separation techniques need long scan time, and are susceptible to physiological motions. Echo planar imaging (EPI) has been adopted for improving the speed of water-fat separation. Nevertheless, the performance of EPI-based water-fat separation is challenged by B0 field inhomogeneity and large fat-shift along the phase-encoding (PE) dimension.2 Consequently, extra scans with different TE shifts are required. As an alternative ultrafast method, spatiotemporally encoded (SPEN)3-4 single-shot MRI possesses better immunity to the B0 field inhomogeneity. Due to special quadratic phase modulation, the SPEN signal holds additional chemical shift information, which can be exploited for the fastest water-fat separation since only single shot is required. Currently, some super-resolved (SR) methods have been proposed for water-fat separation, such as conjugate gradient (CG) method5 and super-resolved water/fat image reconstruction (SWAF) method6. However, the state-of-the art methods still have some shortcomings in spatial resolution, residual artifacts and time consumption. Deep learning is a versatile tool and has been proven successful in many fields recently. In this study, we aimed to propose a deep-learning-based SPEN reconstruction method to obtain water and fat images with high quality simultaneously.Methods
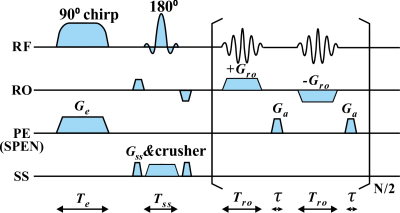
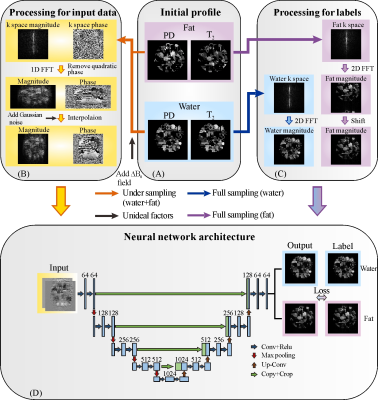
The single-shot SPEN MRI sequence used in this study is shown in Fig. 1. Experiments on in vivo rat abdomen were done on a 7T Varian MRI system (Agilent Technologies, Santa Clara, California), and the fat chemical shift was 1014 Hz in the 7T system. The references were obtained by multi-scan spin echo (SE) sequence with three different echo time shifts and globally optimal surface estimation (GOOSE)7 algorithm. For SPEN MRI, the results obtained by CG and SWAF methods were performed for comparison. The under-sampling rate of SPEN MRI is 50%. Signal-to-ghost ratio (SGR)8 was used to assess the artifact level quantitatively. The higher SGR value means the better artifact suppression. In this study, the training data were generated using synthetic models and MRiLab software9, and this scheme has been successfully validated in our previous studies10. U-Net11 was employed in this study. As illustrated in Fig. 2, the inputs of U-Net are the real and imaginary parts of SPEN signal with quadratic phase removing and signal interpolation, and the outputs are water and fat images.Results and Discussions
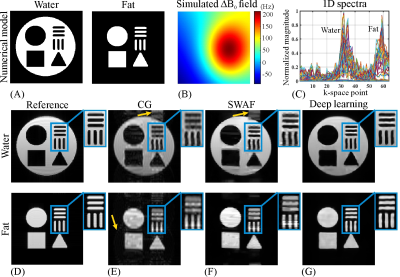
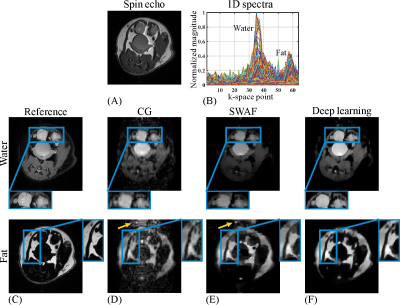
The numerical water/fat model is shown in Fig. 3A. The representative B0 map added in the numerical simulations and the 1D spectra of SPEN signals along the PE dimension are shown in Fig. 3B & C, respectively. The water/fat results reconstructed from different methods are shown in Fig. 3E-G. As indicated by yellow arrows in Fig. 3E & F, the residual artifacts are obvious in the water/fat images obtained by CG and SWAF. On the contrast, these artifacts are indiscernible in deep learning results shown in Fig. 3G. The SGR values of water/fat images reconstructed by CG and SWAF are 16.5dB/23.8dB and 20.9dB/36.2dB, and the SGR value is 35.9dB/41.2dB for deep learning results, which demonstrated the artifact removal ability of our method.The results of in vivo rat experiments are shown in Fig. 4. The anatomical image is shown in Fig. 4A. Similar to the numerical simulations, artifacts caused by incomplete separation are obvious in CG and SWAF results, as indicated by yellow arrows in Fig. 4D & E, while these artifacts are eliminated in deep learning results as shown in Fig. 4F. The SGR values of CG, SWAF, and deep learning results are 19dB/10.3dB, 33.3dB/18.9dB, and 39.3dB/33.8dB for water/fat respectively, which also indicate deep learning can obtain better artifact-suppression performance in both water and fat images. The zoom-in regions indicate that the deep learning method can also obtain sharper edges and better fidelity compared to the other two methods.
Currently, the previous methods still have some challenges. For CG, the ill-conditioned coefficient matrix limits its performance, and for SWAF, filter operation is required for obtaining prior knowledge, which brings loss of details. For the proposed deep learning-based method, the neural network is trained to directly learn the relationship between under-sampled mixed water-fat signal and fully sampled water/fat only signal and no additional filter operation is needed,which guarantees better spatial resolution. As for time consumption, deep learning can obtain separation results much faster than the previous iterative-based methods (e.g. CG and SWAF), especially when dealing with a large number of samples.
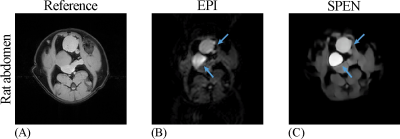
In addition, water-fat separation obtained by single-shot SPEN MRI can also provide competitive results as a fat-suppression EPI strategy when the water-only image is needed. The corresponding results are shown in Fig. 5. As indicated by blue arrows, the separated water images of SPEN are less distorted compared to EPI results. These results imply that SPEN MRI could be a promising ultrafast method to obtain water images with resistance to strong local B0 inhomogeneity and fat contamination, which is a desired property in DWI and many other areas.
Conclusion
SPEN MRI can yield ultrafast water-fat separation within a single shot. In this study, we present a deep learning-based reconstruction method for water-fat separation of SPEN MRI. The proposed method will facilitate the SPEN MRI-based water-fat separation, especially in applications that required high temporal resolution.Acknowledgements
This work is supported by National Natural Science Foundation of China under grant numbers: U1805261, 11775184, and 82071913; Leading (Key) Project of Fujian Province, 2019Y0001.References
1. Eggers H, Bornert P. Chemical shift encoding-based water-fat separation methods. J Magn Reson Imaging. 2014;40(2):251-268.
2. Hu Z, Wang Y, Dong Z, Guo H. Water/fat separation for distortion-free EPI with point spread function encoding. Magn Reson Med. 2019;82(1):251-262.
3. Tal A, Frydman L. Spectroscopic imaging from spatially-encoded single-scan multidimensional MRI data. J Magn Reson. 2007;189(1):46-58.
4. Tal A, Frydman L. Single-scan multidimensional magnetic resonance. Prog Nucl Magn Reson Spectrosc. 2010;57(3):241-292.
5. Schmidt R, Frydman L. In vivo 3D spatial/1D spectral imaging by spatiotemporal encoding: a new single-shot experimental and processing approach. Magn Reson Med. 2013;70(2):382-391.
6. Huang J, Chen L, Chan KWY, Cai C, Cai S, Chen Z. Super-resolved water/fat image reconstruction based on single-shot spatiotemporally encoded MRI. J Magn Reson. 2020;314:106736.
7. Cui C, Wu X, Newell JD, Jacob M. Fat water decomposition using globally optimal surface estimation (GOOSE) algorithm. Magn Reson Med. 2015;73(3):1289-1299.
8. Chen L, Li J, Zhang M, Cai S, Zhang T, Cai C, Chen Z. Super-resolved enhancing and edge deghosting (SEED) for spatiotemporally encoded single-shot MRI. Med Image Anal. 2015;23(1):1-14.
9. Liu F, Velikina JV, Block WF, Kijowski R, Samsonov AA. Fast realistic MRI simulations based on generalized multi-pool exchange tissue model. IEEE Trans Med Imaging. 2017;36(2):527-537.
10. Zhang J, Wu J, Chen S, Zhang Z, Cai S, Cai S, Chen Z. Robust single-shot T2 mapping via multiple overlapping-echo acquisition and deep neural netword. IEEE Trans Med Imaging. 2019;38(8):1801-1811.
11. Ronneberger O, Fischer P, Brox T. U-Net: Convolutional networks for biomedical image segmentation. In: Proceedings of International Intervertion (MICCAI), Munich. Gernamy; 2015;9351:234-241.
Figures