3201
Deep learning based liver segmentation using T1 weighted abdominal MRI1Electrical Engineering, Qatar University, Doha, Qatar, 2Biomedical Engineering and Imaging Institute, Icahn School of Medicine at Mount Sinai, New York, NY, United States, 3Electrical Engineering,, Qatar University, Doha, Qatar, 4Dept. of Physics and Mathematics, North South University, Dhaka, Bangladesh, 5Dept. of Electrical, Electronics and Systems Engineering, Universiti Kebangsaan Malaysia, Bangi, Malaysia, 6Department of Electrical Engineering, University of Engineering and Technology, Peshawar, Pakistan
Synopsis
MR scans are preferred by clinicians for liver pathology diagnosis over volumetric abdominal CT scans, due to their superior resolution for soft tissues. Nevertheless, deep learning based automated liver segmentation from abdominal MRI is challenging as the liver exhibits variable characteristics. This study investigates multiple state-of-the-art segmentation architectures (UNet, UNet++, and FPN) with varying encoder and decoder backbones. Here, T1 weighted MR images are investigated as it demonstrates brighter fat content. Among the investigated networks UNet++ with DenseNet backbone demonstrates top performance for the liver segmentation with a DSC and IoU of 94.3% and 91.0%, respectively.
INTRODUCTION
Automated precise segmentation of the liver is indispensable and a prerequisite for any artificial intelligence (AI) driven liver pathology diagnosis. Nonetheless, such segmentation task is challenging, as liver anatomy can distinctly differ with patients and clinical conditions 1,2. Furthermore, its proximity to adjacent abdominal organs construct a colossal amount of complexity and ambiguity. Magnetic Resonance (MR) imaging is extensively adopted by clinicians for liver pathology investigation, due to their superior contrast and spatial resolution for soft tissues compared to computed tomography (CT) scans 3,4. However, recent researches have demonstrated excellent results for such deep neural network (DNN) driven segmentation task from volumetric abdominal CT scans through utilizing Hounsfield Unit (HoU) scaling for image enhancement 5. Owing to the absence of such homogeneous HoU based enhancement, achieving similar excellence from MR images is challenging. This study investigates the state-of-the-art UNet, UNet++, and Feature Pyramid Network (FPN) segmentation architectures with varying depth of DenseNet encoder-decoder backbones for precise liver segmentation from volumetric T1-weighted (T1w) abdominal MR images. Additionally, fat (and protein) contents in T1w MR images are brighter, henceforth liver is more distinguishable from surroundings. Due to such image characteristics, T1w images provide initially enhanced images for this task and could improve performance.METHODS
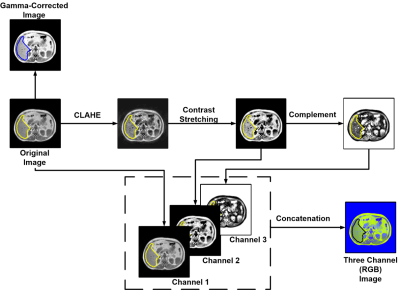
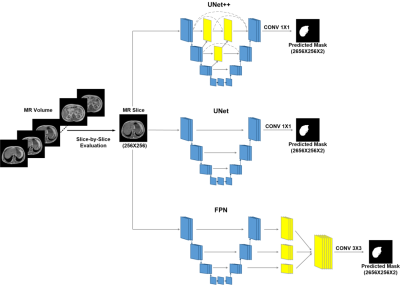
Initially, 20 abdominal T1w MR scans are collected from Combined Healthy Abdominal Organ Segmentation (CHAOS) challenge 6. Ground truth mask of the liver for each MR slices are provided in the dataset, which had been annotated by certified radiologist 6. Each MR scan consists of 26 to 56 slices, and the full dataset holds a total of 647 slices. For the data preparation Matlab is used and deep leanring model development on done in Pytorch using google colabpro. The dataset is split into training, validation, and testing set with a 70:10:20 ratio prior to creating five folds. Affine transformation (rotation and translation) techniques are then implemented on the training set for augmentation. Apart from original images, two image enhancement techniques are investigated. The three-channel RGB images are formed through implementing contrast stretching, adaptive contrast stretching, and image compliment techniques (Fig 1). Gamma-correction is done to have the pixel intensity between 70 to 150 and γ value of 0.5 (Fig 1). Original (non-enhanced) slices, gamma-corrected (contrast stretching) and RGB converted slices are investigated by each of the networks. UNet, UNet++, and FPN segmentation networks with DenseNet-121, DenseNet-160, and DenseNet-201 encoder-decoder backbones are investigated. To take benefit of faster gradient convergence during the training process, pre-trained DenseNets with ImageNet weights are used. In favor of a uniform comparison, training parameters for each of the networks are kept constant (Learning-Rate: 0.0001, Loss-Type: Binary-Cross-Entropy, Optimizer: Adam) (Fig 2).RESULTS
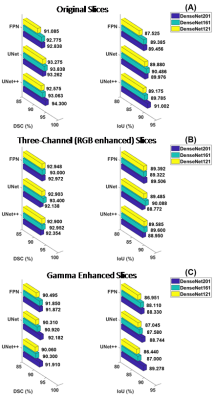
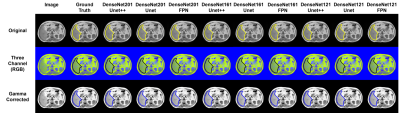
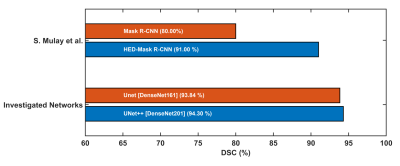
Dice Score (DSC)/F1 Score and Intersection of Union (IoU) are calculated as the performance matrices for the investigated networks. The original slices performed better for each network, compared with the three-channel (RGB) and the gamma-corrected slices. UNet++ with DenseNet201 backbone is the top-performing network when trained on the original slices, provides a DSC and IoU of 94.3% and 91.0%, respectively. On the original slices, UNet with DenseNet161 backbone performed the second best with DSC and IoU of 93.84% and 90.49%. For the three-channel slices, the DSC from UNet++, UNet, and FPN with DenseNet161 backbone are 92.95%, 93.40%, and 93.0%, respectively (Fig. 3). Visualization of the predicted masks illustrates that each of the networks can accurately segment liver boundaries. Additionally, it was observed that the masks predicted from UNet++ with DenseNet201 backbone are smoother in nature. UNet and FPN networks with different backbones also predicted precise masks (Fig 4). S. Mulay et al.5 investigated Holistically-Nested Edge Detection (HED) Mask R-CNN (DSC: 91%) and typical Mask R-CNN (DSC: 80%) for liver segmentation task on the same dataset. The investigated state-of-the-art hybrid architectures outperform such HED-Mask R-CNN based segmentation models by a considerable margin (Fig. 5).DISCUSSION
In this study, the networks performed better on original MR slices compared to the enhanced images. Such a phenomenon illustrates the complexity in implementing image enhancement techniques for such abdominal MR slices. The quality of the contrast stretched and RGB transformed images can be enhanced by strengthening the mask quality and increasing the dataset for training. Further investigation can be conducted focusing on tuning the parameters of contrast stretching and adaptive contrast stretching techniques for this particular case. Moreover, the study is limited within investigating DenseNet based encoder-decoder backbones for the state-of-the-art segmentation architectures. Further investigation with Inception network or Residual network based backbones would enhance performance matrices for the investigated segmentation architectures.CONCLUSION
This study showed that UNet++ with DenseNet backbone demonstrates top performance for the liver segmentation with a DSC and IoU of 94.3% and 91.0%, respectively from original slices of T1w abdominal MRI.Acknowledgements
This research was supported in part by the following grants: NPRP12S-0227-190164 and IRCC-2021-001.References
1. Z. Liu, Y. Song et al. Liver CT sequence segmentation based with improved U-Net and graph cut, 2019, Expert Systems with Applications, 2019, (126): https://doi.org/10.1016/j.eswa.2019.01.055
2. J. Pemg, Y. Want et al. Liver segmentation with constrained convex variational model, Pattern Recognition Letters, 2014, (43): https://doi.org/10.1016/j.patrec.2013.07.010
3. F. C. Alves, J. Brito et al. Liver haemangioma: common and uncommon findings and how to improve the differential diagnosis, European Radiology, 2007, (17): https://doi.org/10.1007/s00330-006-0503-z
4. G. Wang, S. Zhu et al. Comparison of values of CT and MRI imaging in the diagnosis of hepatocellular carcinoma and analysis of prognostic factors, Oncol Lett, 2019, (17); https://doi.org/10.3892/ol.2018.9690 5. K. Kim, J. Chun, A new hyper parameter of hounsfield unit range in liver segmentation, Jour. of Internet Computing and Services, 2020, 21(3); https://doi.org/10.7472/jksii.2020.21.3.103
6. A.E. Kavuar, N.S. Gezer et al. CHAOS Challenge - combined (CT-MR) Healthy Abdominal Organ Segmentation, Medical Image Analysis, 2021, 69: https://doi.org/10.1016/j.media.2020.101950
7. S. Mulay, G. Deepika et al. Liver Segmentation from Multimodal Images Using HED-Mask R-CNN, MMMI, 2019, Lecture Notes in Computer Science, Springer, 11977: https://doi.org/10.1007/978-3-030-37969-89
Figures