3200
WRIST CARTILAGE SEGMENTATION USING U-NET CONVOLUTIONAL NEURAL NETWORKS ENRICHED WITH ATTENTION LAYERS1Faculty of Physics, ITMO University, Saint-Petersburg, Russian Federation, 2Federal Almazov North-West Medical Research Center, Saint-Petersburg, Russian Federation, 3Aix-Marseille Universite, CNRS, Centre de Résonance Magnétique Biologique et Médicale, UMR, Marseille, France
Synopsis
Detection of cartilage loss is crucial for the diagnosis of osteo- and rheumatoid arthritis. An automatic tool for wrist cartilage segmentation may be of high interest as the corresponding manual procedure is tedious. U-Net is a convolution neural network, which has been largely used for biomedical images, but its performance in segmenting wrist cartilage images is modest. Here, we assessed whether adding attention layers to U-Net architecture would improve the segmentation performance. A truncated version of U-Net with attention layers showed the best performance(3D DSC - 0.811), as well as in the accuracy of cartilage cross-section measurements (bias - 6.4 mm2).
INTRODUCTION
Segmentation of MR images of joints to detect cartilage loss is important for the diagnosis of osteo- and rheumatoid arthritis. Although convolutional neural networks (CNNs) have been proved as highly accurate for biomedical image segmentation1, there is still a lack of CNN-based methods for wrist cartilage segmentation. More specifically, previous investigations have shown that U-Net-based segmentation2 could provide good 3D Dice Similarity Coefficient (DSC) against feedforward CNN with standard patch-based architecture, i.e. where a decision on a pixel is made on information from a crop3. In the field of wrist cartilage segmentation, however, the best reported 3D DSC value was still relatively low (0.77) and mainly because of a high occurrence frequency of false-positive results2. In the present work, we investigated whether adding attention layers within U-Net-based CNN's could solve the issue of having false-positive results and increase mean 3D DSC values.METHODS
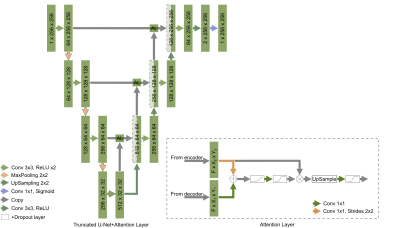
Two basic architectures were used: the classical U-Net and its truncated form. Both CNNs had architectures and parameters similar to what has been previously reported2. Attention layers (AL) have been added to both CNNs and the corresponding performance has been compared4. Feature maps from encoder and decoder paths of U-net act as inputs for each of the attention layers. After size aligning convolutions, feature maps from encoder and decoder paths are element-wise added to each other, so the regions, for which both feature maps have high values of activation functions, are amplified. After several operations, such as ReLU, convolution 1x1, and sigmoid, this procedure provides attention coefficients and the corresponding map is multiplied by the feature map coming from the encoder path. Consequently, the attention layers allow focusing the training process on regions of interest in the feature maps. We hypothesized that this attention procedure may solve the issue of false-positive results outside the joint region. Hyperparameters were optimized in a grid search.The training dataset contained 10 partially labeled 3D MR wrist images resulting in 284 2D images in total2. The training dataset was augmented using the albumentations library and the following types of transformations: vertical and horizontal flip, arbitrary angle rotation, elastic transform, and grid distortion. The dataset was finally normalized using mean and standard deviation values. The final number of 2D images for the augmented dataset was 1520. We used Python 3.6.4, TensorFlow 1.7.0, and Keras 2.1.5. The training and testing procedures were performed using a workstation with GPU Nvidia Tesla V100 a regular PC with GPU Nvidia GTX1050 respectively. The testing procedure was performed using another 10 labeled 3D images i.e. 260 2D images. U-Nets initially trained without attention layers were tested with rotated by 90 degrees data that slightly improve results compared to the previous results2. A set of 20 medial slices was used for estimation of cartilage cross-sectional area (CSA), proposed as a possible metric for cartilage assessment5.
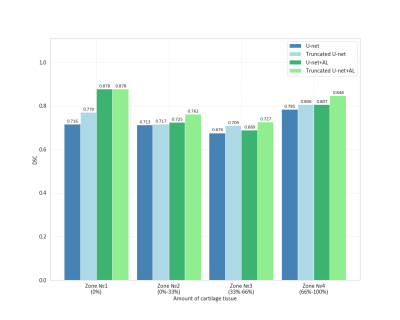
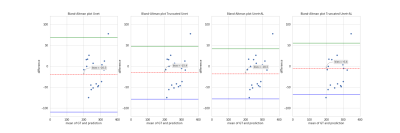
Cartilage masks predicted by CNNs were compared to labels using DSC. We analyzed the distribution of mean 2D DSC values on a layer basis and in areas with a certain amount of cartilage (zone #1 no cartilage, zone #2 from 0 to 33%, zone#3 - from 33% to 66%, and zone #4 – from 66% to 100%). For each CNN, we have computed 3D DSC averaged over 10 3D images from the test dataset. Two additional metrics were used: the area under the precision-recall curve (AUCPR) and the volume overlap error (VOE=(1-IOU)*100%). A Bland-Altman analysis was performed for the cartilage CSA to estimate the performance of the CNNs as automatic tools for CSA measurement. CSA obtained manually and automatically was compared via a paired Student’s t-test computed using R studio.
RESULTS AND DISCUSSION
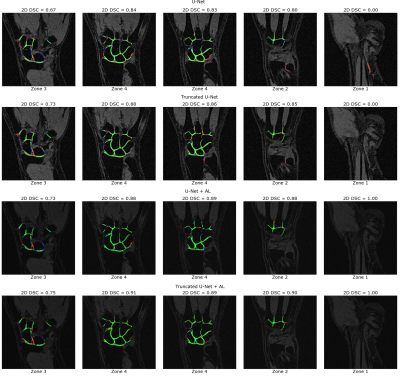
Learning parameters for the U-Net/Truncated U-Net were as follows: batch size – 32, learning rate - 5e-3, noise level -0.3/0.2, dropout probability – 0.4/0.35. These parameters for the corresponding networks with attention layers were: batch size – 32, learning rate - 5e-3/6e-3, noise level - 0.35, dropout probability – 0.3. The structure of an attention layer and the final architecture of U-Net with attention layers are provided in Fig. 1. The results of the zone-based analysis are presented in a bar chart in Fig. 2. An example of networks’ performance in different zones is depicted in Fig. 3. Attention mechanisms significantly improved 2D DSC values in zone #1 (0.878 for both U-Nets + AL). For the other zones, the use of attention layers improved the performance of the truncated U-Net CNN only, with the 3D DSC raising from 0.767 to 0.811. This network also had the largest AUCPR (0.818) and the smallest VOE (31%) values. The corresponding metrics are summarized in a Table in Fig. 4. The Bland-Altman plots presented in Fig.5 indicated that CSA measurements using the truncated U-Net+AL was the most efficient method given that it provided the smallest bias (-6.4 mm2). CSA measurements performed with the truncated U-Net+AL were nonsignificantly different than those performed manually.CONCLUSION
We have investigated the potential of adding attention layers to U-Net architectures regarding the accuracy of wrist cartilage segmentation in 3D MR images. The truncated version of U-Net enriched by attention layers showed the best performance in terms of mean 3D DSC values and accuracy of cartilage CSA measurements.Acknowledgements
This work was supported by the Ministry of Education and Science of the Russian Federation (075-15-2021-592).References
1. Ebrahimkhani, Somayeh. et al. Artificial Intelligence in Medicine, 106, 2020,
2. Brui E.A. et al. Proc. Intl. Soc. Mag. Reson. Med., 2021, Vol. 29, pp. 2969
3. Brui E., et al. NMR in Biomedicine. 2020;33(8):1-13.
4. Ozan Oktay, et al. ArXiv abs/1804.03999 (2018).
5. Zink J.V. et al. World journal of orthopedics. 2015;6(8):641-648
Figures