3199
Adaptive convolution kernels for breast tumor segmentation in multi-parametric MR images1Paul C. Lauterbur Research Center for Biomedical Imaging, Shenzhen Institute of Advanced Technology, Chinese Academy of Sciences, Shenzhen, China
Synopsis
Multi-parametric magnetic resonance imaging (mpMRI) provides high sensitivity and specificity for breast cancer diagnosis. Accurate breast tumor segmentation in mpMRI can help physicians achieve better clinical managements. Existing deep learning models have presented promising performances. However, the effective exploitation and fusion of information provided in mpMRI still need further investigation. In this study, we propose a convolutional neural network (CNN) with adaptive convolution kernels (AdaCNN) to automatically extract and absorb the useful information from multiple MRI sequences. Extensive experiments are conducted, and the proposed method can generate better breast tumor segmentation results than those obtained by CNNs with normal convolutions.
Introduction
Breast cancer persists to be the most common cancer type diagnosed in women. In 2021, breast cancer accounts for 30% of all female cancers [1]. Early detection and treatment are crucial for the survival of breast cancer patients [2]. Magnetic resonance imaging (MRI) is an important tool in the clinic for breast cancer screening. Multi-parametric MRI (mpMRI) combining different imaging sequences (such as contrast-enhanced MRI (CE-MRI) and T2-weighted MRI) provides high sensitivity and specificity [3-5]. Accordingly, automated methods, which can process mpMRI data, are needed to assist physicians in achieving fast and accurate breast cancer detection [6]. Existing studies suggest that special network architectures or modules are needed to effectively fuse the information provided by multi-modal inputs or mpMRI data [7,8]. However, building such dedicated network components is difficult and cumbersome in the first place. Moreover, misalignment between mpMRI can happen, which may bring new issues to the segmentation task.In this study, we propose to automatically extract useful information from breast mpMRI without any specifically designed modules. No alignment between different MRI sequences is required. Particularly, inspired by conditional convolutions [9,10], we design a CNN with adaptive convolution kernels (AdaCNN) that can adaptively learn the useful information from MR images acquired with different scan parameters. Once the mpMRI information is absorbed by the network parameters, the model can be tested using MR data from a single sequence. With AdaCNN, better results are obtained with CE-MRI inputs compared to those achieved by baseline models with either single sequence MR data or multi-parametric data.
Methodology
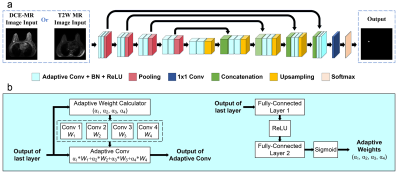
The overall workflow of our proposed breast tumor segmentation framework is depicted in Figure 1. The underline network architecture adopts a classical encoder-decoder structure. All convolutions except the last convolution with a kernel size of 1x1 for pixel-wise classification are replaced by adaptive convolutions, which are introduced to adaptively adjust the convolution kernels according to the inputted image data. Specifically, a normal convolution is divided into K convolutions (we set K = 4) with parameters of (W1, W2, …, WK), which can be treated as K experts. Input-dependent adaptive weights are calculated in a way similar to the attention module [11]. With the calculated weights, the experts are combined linearly to form a new adaptive convolution. The parameters of the adaptive convolution change with the inputs, and thus, it can implicitly learn the difference between the MRI data acquired with different scanning parameters and adaptively modify the kernels to better process the inputs.Two baseline models are investigated, and the results are reported to validate the effectiveness of the proposed method. One baseline (UNet) has the same network as the proposed method except that the convolutions are normal convolutions [12]. The other baseline (FuseNet) explicitly fuses the features extracted from mpMRI data in the network and requires multi-parametric inputs during testing [8]. Experiments are conducted with an in-house dataset with approval from the local ethics committee. Breast MRI scans using two MR sequences (contrast-enhanced T1-weighted MRI (T1CE) and T2-weighted MRI (T2W)) were collected. All images were acquired using a Philips Achieva 1.5T system with a four-channel phased-array breast coil. In total, data from 313 patients are collected. Five-fold cross-validation experiments are conducted. Each experiment is replicated three times. Results are presented with mean ± s.d.
Results and Discussion
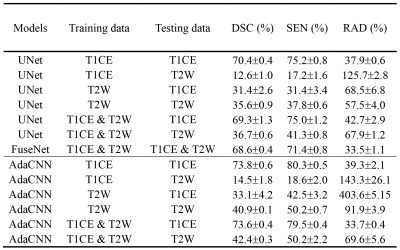
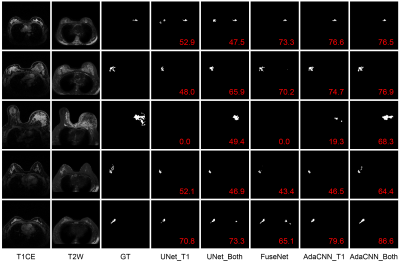
Three evaluation metrics are calculated and reported, including Dice similarity coefficient (DSC), sensitivity (SEN), and relative area difference (RAD). Results are listed in Table 1. It can be observed that compared to the baseline UNet, FuseNet gets even worse performance due to the ineffective fusion of information provided in the multi-parametric breast MR data. On the other hand, the proposed AdaCNN can appropriately explore the information of the respective MR sequence data. Better results are achieved by AdaCNN compared to the baseline CNNs. For AdaCNN, when it is trained with the combined dataset, it can generate the best segmentation results for both sequences with a single model (last two rows in Table 1). Figure 2 plots several example segmentation maps of different models, which can lead to the same conclusion as the quantitative results.Conclusion
In this study, a CNN model with adaptive convolutions is proposed to automatically learn the useful information provided in breast mpMRI data. The proposed model, which is optimized with mpMRI data, can generate the best segmentation results during testing without requiring mpMRI data when compared to the baseline models tested with either MRI data from a single sequence or multiple sequences. Therefore, the proposed method is more applicable to real-life clinical scenarios.Acknowledgements
This work was partly supported by Scientific and Technical Innovation 2030-“New Generation Artificial Intelligence” Project (2020AAA0104100, 2020AAA0104105), the National Natural Science Foundation of China (61871371, 81830056), the Basic Research Program of Shenzhen (JCYJ20180507182400762), and Youth Innovation Promotion Association Program of Chinese Academy of Sciences (2019351).References
[1] Siegel, R. L., Miller, K. D., Fuchs, H. E. & Jemal, A. Cancer statistics, 2021. CA. Cancer J. Clin. 71, 7–33 (2021).
[2] Barber, M. D., Jack, W. & Dixon, J. M. Diagnostic delay in breast cancer. Br. J. Surg. 91, 49–53 (2004).
[3] Liang, C. et al. An MRI-based radiomics classifier for preoperative prediction of Ki-67 status in breast cancer. Acad. Radiol. 25, 1111–1117 (2018).
[4] Westra, C., Dialani, V., Mehta, T. S. & Eisenberg, R. L. Using T2-weighted sequences to more accurately characterize breast masses seen on MRI. Am. J. Roentgenol. 202, 183–190 (2014).
[5] Marino, M. A., Helbich, T., Baltzer, P. & Pinker-Domenig, K. Multiparametric MRI of the breast: A review. J. Magn. Reson. Imaging 47, 301–315 (2018).
[6] Hu, Q., Whitney, H. M. & Giger, M. L. Radiomics methodology for breast cancer diagnosis using multiparametric magnetic resonance imaging. J. Med. Imaging 7, 1–15 (2020).
[7] Li, C. et al. Learning cross-modal deep representations for multi-modal MR image segmentation. In International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI) 11765 LNCS, 57–65 (2019).
[8] Hazirbas, C. & Ma, L. FuseNet: Incorporating depth into semantic segmentation via fusion-based CNN architecture. In Asian Conference on Computer Vision (ACCV) (2016).
[9] Yang, B., Le, Q. V., Bender, G. & Ngiam, J. CondConv: Conditionally parameterized convolutions for efficient inference. In Advances in Neural Information Processing Systems (NeurIPS) 32, (2019).
[10] Chen, Y. et al. Dynamic convolution: Attention over convolution kernels. In IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 11027–11036 (2020).
[11] Hu, J., Shen, L. & Sun, G. Squeeze-and-excitation networks. In IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 7132–7141 (2018).
[12] Falk, T. et al. U-Net: deep learning for cell counting, detection, and morphometry. Nat. Methods 16, 67–70 (2019).
Figures