3198
Unsupervised Domain Adaptation via CycleGAN for knee joint Segmentation in MR Images1The Chinese University of Hong Kong, Hong Kong, Hong Kong, 2The City University of Hong Kong, Hong Kong, Hong Kong
Synopsis
Knee joint tissues segmentation is necessary for quantitative analysis of musculoskeletal diseases like knee osteoarthritis. Three-dimensional Fast Spin Echo (3D FSE) imaging is a potential MRI technique for routine clinical knee imaging. Thus, segmentation based on 3D FSE has valuable clinical application. However, the conventional deep learning-based segmentation requires manually annotating 3D knee images which is time-consuming. In this work, we proposed a domain adaption-based unsupervised approach for cartilage and meniscus segmentation on 3D FSE images without the need for annotating images. We demonstrated that the proposed method improved the quality of segmentation.
Introduction
Knee osteoarthritis (OA) is a prevalent degenerative musculoskeletal disease worldwide1. Accurate morphology analysis of articular cartilages and meniscus based on MRI can be used to characterize knee OA. Automatic segmentation of knee tissues is essential to provide such analysis in routine clinical practice. Fast Spin Echo (FSE) imaging plays a central role in knee MRI. Three-dimensional(3D) acquisition of FSE can be used to acquire images with isotropic resolution and reformatted to arbitrary plane to reduce total acquisition time2. Thus, automatic knee joint tissue segmentation based on 3D FSE acquisition has valuable clinical applications. It is time-consuming and prone to inter and intrareader variance to create a labeled dataset of 3D knee MRI1. Unsupervised Domain Adaptation (UDA) has been investigated to transfer feature representations learned from the source data domain to the target domain3. In this work, we propose a deep learning-based UDA approach for knee joint segmentation of unannotated 3D FSE images. The proposed approach uses a publicly available annotated dataset from the Osteoarthritis Initiative (OAI), two U-Nets4 for segmentation, and one Cycle‐consistent Generative Adversarial Network (CycleGAN)5 for image translation between different MR imaging sequences.Methods
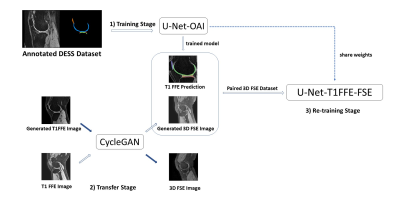
The study was conducted under the approval of the institutional review board. The data sets used in this study include A) 88 data sets acquired using 3D DESS acquisition with segmentation labels from the OAI project; B) 13 3D T1 weighted Fast Field Echo (FFE) images (resolution 0.5mm x 0.5mm x 1mm); This data set had no segmentation labels and served as an intermediate imaging sequence; C) 13 proton density-weighted 3D FSE images acquired using commercial pulse sequence VISTATM (Philips Healthcare) (resolution 0.8mm x 0.8mm x 0.8mm). Both data set B and C were acquired at a 3.0T Philips Achieva TX scanner (Philips Healthcare) using an 8-channel knee coil. Data set B and C are paired as these datasets are acquired from the same patients. Manual segmentation was carried out to obtain segmentation labels of data set C by a musculoskeletal radiologist using ITK-SNAP. These segmentation labels are used as the ground truth to validate the proposed method for the segmentation of 3D FSE images. The segmentation labels include 6 classes, femoral cartilage (FC), medial meniscus (MM), medial tibial cartilage (MTC), lateral meniscus (LM), lateral tibial cartilage (LTC), patellar cartilage (PC), and background.Figure 1 shows the pipeline of our proposed method for cartilage and meniscus segmentation. Our method consists of the 1) training stage, the 2) transfer stage, and the 3) re-training stage. First, at the 1) training stage, a U-Net with an Efficientnet6 encoder, U-Net-OAI, is trained using the large, annotated dataset from OAI (data set A). This model is used to generate segmentation labels for T1 FFE images (data set B). We observed reliable predictions can be generated from U-Net-OAI. At the 2) transfer stage, a CycleGAN performs a domain adaptation from T1 FFE images to 3D FSE images. The CycleGAN is trained on the T1 FFE images and 3D FSE images to synthesize 3D FSE visual contrast from the T1 FFE images. The precise adaption was achieved as both the T1 FFE image and the 3D FSE image were acquired from the same patients. At 3) re-training stage, the data used is the generated paired 3D FSE data sets consisting of synthesized 3D FSE images from corresponding T1 FFE images, and T1 FFE labels from U-Net-OAI. In this stage, the U-Net-OAI pre-trained on 1) can be fine-tuned to learn feature representations between 3D FSE images and 3D FSE labels, to get the final model, U-Net-T1FFE-FSE.
In addition to the above method, we prepared another CycleGAN trained by OAI and 3D FSE data for comparison. In this scenario, the model, U-Net-FSE, fine-tuned in the 3) stage is trained by OAI labels and the synthesized 3D FSE images.
Results
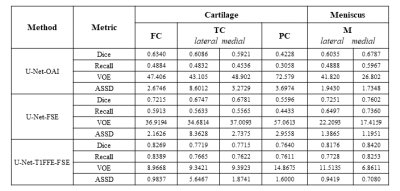
In our study, we evaluated the result of the proposed approach. The result of U-Net-OAI, U-Net-FSE, and U-Net-T1FFE-FSE achieved the average DICE of 0.590, 0.687, and 0.799 respectively. The scores of Dice coefficient (DICE), Recall, Volumetric Overlap Error (VOE), and Average Symmetric Surface Distance (ASSD) on each tissue are shown in Figure 2. By the unsupervised domain adaption, both U-Net-FSE and U-Net-T1FFE-FSE improved segmentation performance at all metrics. Furthermore, U-Net-T1FFE-FSE performed the most accurately in all knee tissues. Compared to the U-Net-OAI model, the U-Net-T1FFE-FSE improves the segmentation performance on 3D FSE images from 0.634 to 0.722 for FC, 0.609 to 0.675 for LTC, 0.592 to 0.678 for MTC, 0.423 to 0.764 for PC, 0.604 to 0.725 for LM and 0.679 to 0.760 for MM respectively.Discussion and Conclusions
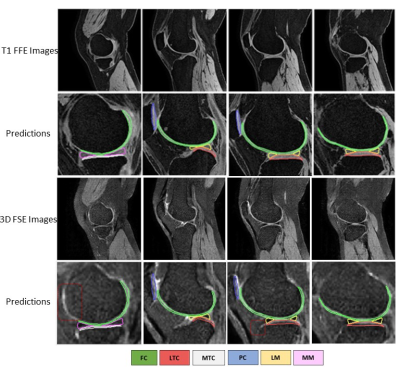
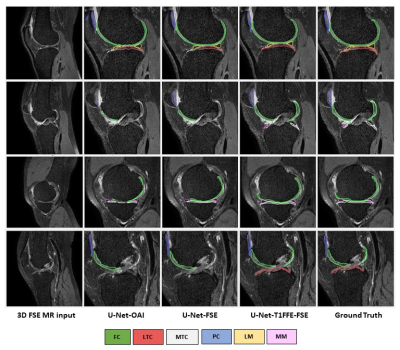
In our study, the leverage of generated pair 3D FSE (VISTA) data greatly improves the segmentation predictions of 3D FSE MRI, particularly in patella cartilage. The U-Net-OAI model has inferior performance on segmentation of 3D FSE images compared to T1FFE images (Figure 3), likely due to a more significant domain shift problem between the OAI data and VISTA. By using the proposed UDA, significant improvement was achieved to segmenting 3D FSE images without the need of annotating them (Figure 4). Further work includes a comparison of the proposed method with other unsupervised networks and its generalization to other applications.Acknowledgements
This study was supported by a grant from the Innovation and Technology Commission of the Hong Kong SAR (Project MRP/001/18X), and a grant from the Research Grants Council of the Hong Kong SAR (Project SEG CUHK02).References
1. Ebrahimkhani, Somayeh, et al. "A review on segmentation of knee articular cartilage: from conventional methods towards deep learning." Artificial Intelligence in Medicine 106 (2020): 101851.
2. Gold, Garry E., et al. "Isotropic MRI of the knee with 3D fast spin-echo extended echo-train acquisition (XETA): initial experience." American Journal of Roentgenology 188.5 (2007): 1287-1293.
3. Toldo, Marco, et al. "Unsupervised domain adaptation in semantic segmentation: a review." Technologies 8.2 (2020): 35.
4.Ronneberger, Olaf, Philipp Fischer, and Thomas Brox. "U-net: Convolutional networks for biomedical image segmentation." International Conference on Medical image computing and computer-assisted intervention. Springer, Cham, 2015.
5. Zhu, Jun-Yan, et al. "Unpaired image-to-image translation using cycle-consistent adversarial networks." Proceedings of the IEEE international conference on computer vision. 2017.
6. Tan, Mingxing, and Quoc Le. "Efficientnet: Rethinking model scaling for convolutional neural networks." International Conference on Machine Learning. PMLR, 2019.
Figures