3190
Deep Learning-based Generative Adversarial Registration NETwork (GARNET) for Hepatocellular Carcinoma Segmentation: Multi-center Study1Institute of Science and Technology for Brain-Inspired Intelligence, Fudan University, Shanghai, China, 2Philips Healthcare, Shanghai, China, 3Department of Radiology, Ruijin Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, China, 4Shanghai Universal cloud imaging dignostic center, Shanghai, China, 5Human Phenome Institute, Fudan University, ShangHai, China
Synopsis
This study proposed a deep learning-based generative adversarial registration network (GARNET) for multi-contrast liver image registration and evaluated its value for hepatocellular carcinoma (HCC) segmentation. We used generative adversarial net (GAN) to synthesize images from diffusion-weighted imaging (DWI) to dynamic contrast-enhanced (DCE) and then applied for deformable registration on the synthesized DCE images. A total of 607 cirrhosis patients from 5 centers (401 HCC patients) were included in this study. We compared the proposed method with symmetric image normalization (SyN) registration and VoxelMorph. Experimental results demonstrated that GARNET improved the registration performances significantly and yielded better segmentation of HCC lesions.
Introduction
Accurate hepatocellular carcinoma (HCC) lesion detection and segmentation rely heavily on accurate registration among different contrast-weighted images, which is challenging due to the large variability of signal intensity distribution among multi-contrast images and the spatial misalignment caused by non-rigid deformations. Current available landmark-based1-4 or intensity-based5-8 methods have a greater probability of registration failure. Hence this study proposed a deep learning-based generative adversarial registration network (GARNET) to improve the registration accuracy of HCC between abdominal diffusion-weighted imaging (DWI) and dynamic contrast-enhanced (DCE) images, and then validated its value in HCC lesion segmentation using attention-based U-net (AU-NET).Methods
Data AcquisitionThis retrospective study included 607 patients with liver cirrhosis (401 cases with HCC) from Jan. 2013 to Feb. 2021. MR imaging data were obtained from 5 medical centers (termed as RHP, PHP, PHS, ZYG, NHG) and four manufacture vendors, 3.0-T Skyra (Siemens), 3.0-T Ingenia (Philips), 1.5-T SIGNA HDe (GE), and 3.0-T Signa HDxt (GE). Two sequences were performed: (a) DWI (reconstructed resolution, 1.56×1.56×10 mm; imaging matrix, 256×256; and b-values, 0 and 600/800/1000 s/mm2), (b) four dynamic phases DCE (reconstructed resolution, 0.78×0.78×3.5 mm; and imaging matrix, 512×512). The delayed phase DCE was chosen as the target image for registration. The target HCC lesions and liver contours were manually outlined by two experienced radiologists on both DWI and delayed phase DCE images independently.
GARNET Registration
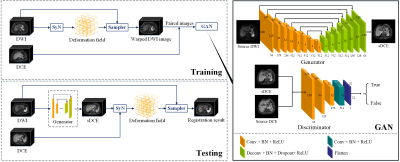
As shown in Figure 1, the proposed GARNET contained a training and a testing stage. We registered DWI and the delayed phase DCE images during the training stage by SyN9, which maximized the cross-correlation within the space of diffeomorphic maps. The training of GAN was performed on 2D slices of 206 non-HCC patients in RHP center with Adam optimizer. The pre-trained GAN was used during the registration stage to generate a synthesized DCE (sDCE) image from the source DWI image. Next, SyN was used to register the sDCE images with source DCE. Then, the deformation field obtained by SyN was applied to the source DWI for final registration.
GAN Architecture
The architectures of the generator and discriminator are shown in Figure 1. A U-Net architecture10 with a symmetrical encoding-decoding structure was used as the generator, containing a contracting path, a symmetric expanding path, and concatenations between the corresponding layers in contracting and expanding path layers. The discriminator network used the patchGANs11 structure, which classified each patch in one image as real or fake. Thus, the discriminator focused on the high-frequency information from the local image patches.
Validation of HCC Segmentation
An attention-based 2.5D U-Net (AU-NET) were used for lesion segmentation. The architecture of AU-NET consisted of two down-sampling and two up-sampling steps and attention-based squeeze-and-excitation blocks. The feature maps of the start, bottom, and final layers were concatenated to fuse multi-level information. We trained an AU-NET with registered DWI (high-b-value) from GARNET, source DCE image, and DCE tumor label as multi-channel input, named as ‘registered AU-NET.’ Compared with AU-NET trained with source DWI (‘unregistered AU-NET’), ‘registered AU-NET’ performed better lesion segmentation results. The output of AU-NET was predicted binary tumor label after connected component denoising, which eliminated noisy predicted label. These two networks were tested in PHP, PHS and NHG centers with 127 cases (ZYG datasets were used for training networks).
Registration and Segmentation Evaluations
In registration and segmentation tasks, dice similarity coefficient (DSC), target registration error (TRE), and Hausdorff distance (HD)12 were used to evaluate the overlap-based and distance-based similarity between two binary masks A and B, and TRE was defined as the distance between the barycenter of binary mask after registration.
Results
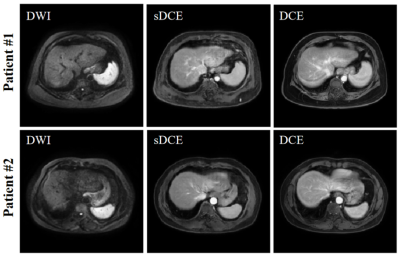
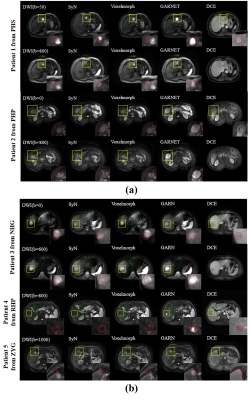
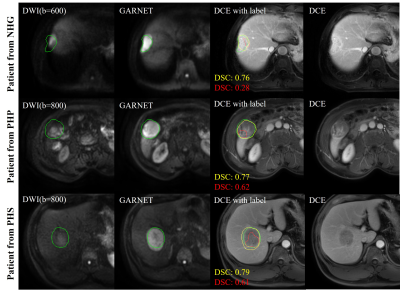
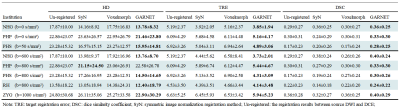
The performance of GAN is shown in Figure 2. The liver contour and vessels in DWI became more obvious in the synthesized DCE image, which was of great help for the subsequent registration. We selected patients from each center with different scan parameters of multi-vendor settings to demonstrate the performances of GARNET. As shown in Figure 3, Compared with SyN and VoxelMorph, results from GARNET were more consistent with the ground truth in DCE in all five centers. In quantitative registration evaluation, our method performed the best in NH and PHS centers. TRE was decreased from 6.92 ± 3.26 to 4.31 ± 3.09 in PHS b-800 group and 4.89 ± 3.06 in PHS b0 group; 5.19 ± 2.37 to 3.73 ± 2.01 in NH b-600 group and 3.85 ± 1.94 in NH b0 group, respectively. The detailed results are shown in Table 1.In segmentation task, the AU-NET with registered multi-contrast images was significantly superior to unregistered AU-NET in three centers in terms of DSC from 0.44 ± 0.26 to 0.56 ± 0.19 in PHS center; from 0.53 ± 0.25 to 0.58 ± 0.21 in PHP center and from 0.36 ± 0.22 to 0.45 ± 0.22 in NGH center. Three cases were selected from the above centers (Figure 4).Discussion and Conclusion
This study proposed a GARNET method to improve the registration accuracy between liver DWI and DCE images. The results proved that GARNET outperformed other methods in terms of higher DSC and lower TRE/HD. In addition, we demonstrated that accurate registration significantly improved the performance of HCC lesion segmentation using an AU-NET. The generalization performance was verified on multi-center datasets.Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 81971583), National Key R&D Program of China (No. 2018YFC1312900), Shanghai Natural Science Foundation (No. 20ZR1406400), Shanghai Municipal Science and Technology Major Project (No.2017SHZDZX01, No.2018SHZDZX01) and ZJLab.References
1. Shen, D,Davatzikos C, HAMMER: hierarchical attribute matching mechanism for elastic registration. IEEE transactions on medical imaging, 2002. 21(11): p. 1421-1439.2. Yang, J, Li H,Jia Y. Go-icp: Solving 3d registration efficiently and globally optimally. 2013 IEEE International Conference on Computer Vision.
3. Liu, Y, Foteinos P, Chernikov A, et al., Mesh deformation-based multi-tissue mesh generation for brain images. Engineering with Computers, 2012. 28(4): p. 305-318.
4. Jian, B,Vemuri BCJITPAMI, Robust Point Set Registration Using Gaussian Mixture Models. IEEE transactions on pattern analysis and machine intelligence 2011. 33(8): p. 1633-1645.
5. Hassan, Rivaz, Sean, et al., Automatic Deformable MR-Ultrasound Registration for Image-Guided Neurosurgery. IEEE transactions on medical imaging 2015.
6. Klein, S, Van dH, Uulke A., Lips IM, et al., Automatic segmentation of the prostate in 3D MR images by atlas matching using localized mutual information. Medical Physics 2008. 35(4): p. 1407-1417.
7. Rivaz, H, Karimaghaloo Z, Fonov VS, et al., Nonrigid Registration of Ultrasound and MRI Using Contextual Conditioned Mutual Information. IEEE transactions on medical imaging2014. 33(3): p. 708-725.
8. Staring, M, Van dH, U. A., Klein S, et al., Registration of Cervical MRI Using Multifeature Mutual Information. IEEE Transactions on Medical Imaging 2009. 28(9): p. p.1412-1421.
9. Avants, BB, Epstein CL, Grossman M, et al., Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Medical image analysis, 2008. 12(1): p. 26-41.
10. Ronneberger, O, Fischer P,Brox T. U-Net: Convolutional Networks for Biomedical Image Segmentation. in medical image computing and computer assisted intervention. 2015.
11. Isola, P, Zhu J, Zhou T, et al. Image-to-Image Translation with Conditional Adversarial Networks. 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). 2017.
12. Huttenlocher, DP, Klanderman GA, Rucklidge WJJITopa, et al., Comparing images using the Hausdorff distance. , IEEE transactions on pattern analysis and machine intelligence 1993. 15(9): p. 850-863.
Figures