3188
Deep Learning Based Automated Diagnosis of Epilepsy in Patients with WHO II-IV Grade Cerebral Gliomas from Multiparametric MRI1Shanghai Key Laboratory of Magnetic Resonance, East China Normal University, Shanghai, China, 2Department of MRI, The First Affiliated Hospital of Zhengzhou University, Zhengzhou, China, 3MR Scientific Marketing, Siemens Healthineers, Shanghai, China
Synopsis
We had retrospectively enrolled 371 glioma patients in this study to develop an automated scheme to predict epilepsy in patients with WHO II-IV grade cerebral gliomas from multi-parametric MRI (mp-MRI). Gliomas tumor was segmented by a segmentation model trained with nnU-Net. Then a classification model based on ResNet-18 using segmented tumor region as anatomical attention was used to predict epilepsy from mp-MRI images. In the independent test cohort, the segmentation model achieved a mean dice of 0.899, while the classification model achieved an AUC of 0.890, better than the baseline ResNet-18 model with a test AUC of 0.783.
Introduction
The cerebral gliomas are the most common primary tumors in adults. Half of all glioma patients and up to 89% low grade glioma (LGGs) patients have epilepsy experience1-2. Glioma-associated epilepsy (GAE) greatly impairs the patients’ life quality, and the patients could develop status epilepticus and multiple seizures without regaining consciousness. Early diagnosis of epilepsy is vital for protecting neurocognitive function, limiting progression and improving patients’ quality of life.Multi-parametric magnetic resonance imaging (mp-MRI) is an essential preoperative examination for patients with glioma3 and convolutional neural networks (CNNs) have been used in glioma-related tasks, such as tumor segmentation and classification4-5. Thus, in this study, we aimed to develop a fully-automated approach to predict epilepsy in patients with WHO II-IV grade cerebral gliomas, with a segmentation model to outline gliomas tumor and an attention-guided classification model for epilepsy diagnosis.
Methods
We had enrolled 371 consecutive glioma patients who underwent MRI scanning before surgery at the First Affiliated Hospital of Zhengzhou University from August 2016 through August 2019. MR images of patients were scanned on 3T MRI scanners (Magnetom Trio TIM/Prisma, Verio or Skyra, Siemens Healthcare; Discovery 750, GE Medical Systems). The dataset was divided into 298 (163 epilepsy/135 no epilepsy) training cohort and 73 (40 epilepsy/33 no epilepsy) test cohort. For each patient, T2-weighted (T2W) images, apparent diffusion coefficient (ADC) maps (calculated with diffusion-weighted images b=0, 1000s/mm2) and post-contrast T1-weighted (T1Gd) images were aligned onto T2 Fluid Attenuated Inversion Recovery (T2-FLAIR) images with Elastix. All images were resampled to a resolution of 0.688mm×0.688mm×6.500mm (inter-slice) and padded to 360×360×20. Volumes of interest (VOIs), including the entire tumor and peritumoral edema, were manually drawn slice-by-slice on T2-FLAIR images by a radiologist with 5 years of experience in neuro-oncology and were reviewed and modified (if necessary) by a neuroradiologist with 20 years of experience in neuroradiology.A modified nnU-Net6 based on T2-FLAIR images was trained to accomplish automated segmentation of the tumor and peritumoral edema. The nnU-Net framework is a convenient and effective segmentation method designed to deal with diverse dataset. We used 5-fold cross validation to train the model, using stochastic gradient descent (SGD) as optimizer, with a learning rate of 0.01, momentum of 0.99, and the max epoch of 300.
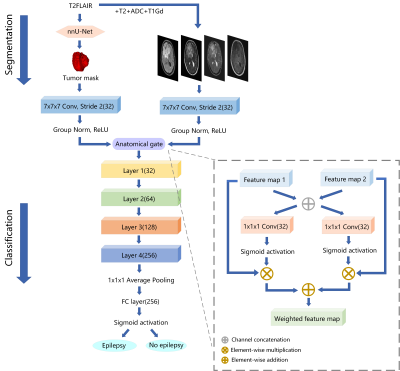
Our classifier for epilepsy prediction was derived from the well-known 18-layer ResNet architecture and a novel anatomical attention gate7 (Figure 1). 3D T2-FLAIR, T2W, T1Gd images, and ADC maps were concatenated in the channel dimension. Then the multi-channel image and tumor segmentation were parallelly fed into 7×7×7 convolution and through an anatomical gate to fuse feature maps generated by MR images and tumor region. The proposed model contained four residual blocks and each block included two 3×3×3 convolutional layers follow by BN and ReLU. We used Adam as optimizer, with an initial learning rate of 0.001. Prediction of epilepsy was mean ensembled by five models selected through a 5-fold cross validation.
We compared the performance of proposed classifier with the baseline ResNet-18, which only used multi-channel images as input. The receiver operating characteristic (ROC) analysis and confusion matrix were utilized to evaluate the classification performance.
Results
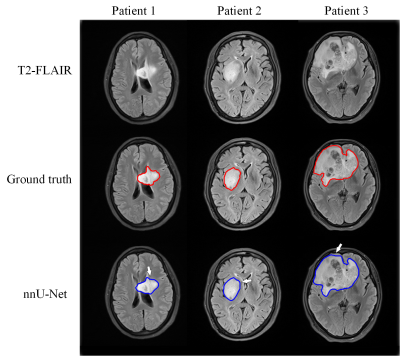
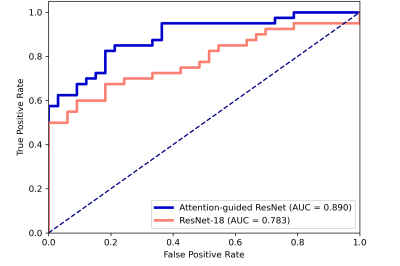
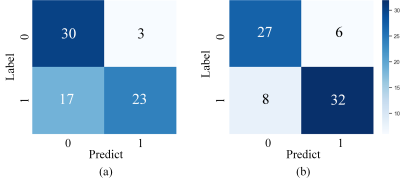
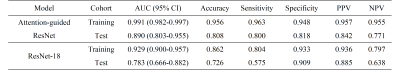
The segmentation model achieved a mean dice of 0.890±0.118 and 0.899±0.064 in the training and independent test cohort, respectively. Figure 2 shows the segmentation results and ground truth from three patients in the test cohort.Our attention-guided classification model achieved a training AUC of 0.991 (95% confidence interval (CI): 0.982-0.997) and a test AUC of 0.890 (95% CI: 0.803-0.955), while baseline model obtained a training AUC of 0.929 (95% CI: 0.900-0.957) and a test AUC of 0.783 (95% CI: 0.666-0.882). The ROC curves of our attention-guided model and baseline model were plotted in Figure 3. The confusion matrix of two models were shown in Figure 4 and statistics analysis was listed in Table 1.
Discussion and Conclusion
In this study, we proposed a fully automated approach to diagnose epilepsy for patients with different grade glioma from mp-MRI, with a segmentation model and a classification model. Manual segmentation requires a lot of expertise and skills, while automatic segmentation reduced workload and improved consistency. Although our segmentation was not perfect, it could still help to improve diagnosis via attention mechanism. The anatomical attention gate fusing glioma tumor region and mp-MRI feature maps can be employed as the guidance to improve the prediction. Hence, our method outperformed baseline ResNet-18 in the prediction of epilepsy with a higher AUC and accuracy on the same test cohort. Compared with previous work8, this study covers not only LGG but also high grade of glioma with a better performance in diagnosis of epilepsy.Since the preoperative diagnosis of GAE was made based on clinical signs, electroencephalography (EEG) and imaging findings, a neuroradiologist is hard to diagnosis epilepsy only on MRI9. However, our work showed that mp-MRI has the potential to diagnosis epilepsy without other information, making diagnosis a non-invasive, automated and easy-to-use process. In the future, this study will be extended to incorporate radiomics feature to the deep learning classification model, and software integrating the whole pipeline will be developed so that the approach can be test further in a multi-institutional, clinical environment.
Acknowledgements
This project is supported by National Natural Science Foundation of China (61731009).References
1. Englot DJ, Chang EF, Vecht CJ. Epilepsy and brain tumors. Handb Clin Neurol. 2016; 134: 267–85.
2. Pallud J, Audureau E, Blonski M, et al. Epileptic seizures in diffuse low-grade gliomas in adults. Brain. 2014; 137(2): 449–462.
3. Upadhyay N, Waldman AD. Conventional MRI evaluation of gliomas. Br J Radiol. 2014; 84(2): 79-226.
4. Korfiatis P, Erickson B. Deep learning can see the unseeable: predicting molecular markers from MRI of brain gliomas. Clin Radiol. 2019; 74(5): 367-373.
5. Naser MA, Deen MJ. Brain tumor segmentation and grading of lower-grade glioma using deep learning in MRI images. Comput Biol Med. 2020; 121.
6. Isensee F, Jaeger PF, Kohl SA, et al. nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nat Methods. 2020; 18(2): 203–211.
7. Sun L, Shao W, Zhang D, et al. Anatomical Attention Guided Deep Networks for ROI Segmentation of Brain MR Images. IEEE Trans Med Imaging. 2020; 39(6): 2000-2012.
8. Liu Z, Wang Y, Liu X, et al. Radiomics analysis allows for precise prediction of epilepsy in patients with low-grade gliomas. Neuroimage Clin. 2018; 19: 271–278.
9. Liang S, Fan X, Zhao M, et al. Clinical practice guidelines for the diagnosis and treatment of adult diffuse glioma-related epilepsy. Cancer Med. 2019; 8(10): 4527-4535.
Figures