3184
3D Automatic segmentation of breast lesion in dynamic contrast enhanced MRI using deep convolutional neural network1Paul C. Lauterbur Research Center for Biomedical Imaging, Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, Shenzhen, China, 2Zhengzhou University, Zhengzhou, China, 3Department of Radiology, National Cancer Center/National Clinical Research Center for Cancer/Cancer Hospital & Shenzhen Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Shenzhen, China, 4Henan University, Kaifeng, China
Synopsis
Breast cancer is the most common cancer in women with the highest incidence. Dynamic contrast enhanced MRI is one of the backbone sequences for breast cancer diagnosis. Accurate segmentation of breast lesions based on DCE-MRI images is helpful for clinically objective and quantitative evaluation of breast lesions. However, the commonly used manual segmentation method is subject to high inter-observer variability. In this study, a 3D automatic algorithm is proposed for segmentation of breast lesions in DCE-MRI. The results show that the proposed network can obtain accurate and automatic 3D segmentation of breast lesions and achieves better segmentation results than VNet.
INTRODUCTION
At present, breast cancer has become one of the most common threats to women's physical and mental health worldwide. Statistics showed that the incidence rate of breast cancer in women reached 24.2% with the mortality rate of as high as 15%., which is the highest incidence and mortality rate among female cancers [1]. In all sequences of routine breast MRI, DCE-MRI is a major sequence, not only because it is the most sensitive method of detecting breast cancer [2], but more importantly, it can reflect the heterogeneity of tumor hemodynamics, including micro-vessel density and vascular permeability difference [3]. Therefore, it is very important to accurately segment breast lesions from DCE images, which is very helpful for clinically objective and accurate evaluation of the hemodynamic heterogeneity of breast lesions. However, the commonly used manual segmentation methods are time-consuming and labor-intensive, and rely heavily on the subjective experience of doctors, especially for 3D segmentation of lesions, thus subject to inter-observer variability. In this work, a network structure based on cascaded v-net is proposed for automatic segmentation of 3D breast lesions on DCE-MRI.METHODS
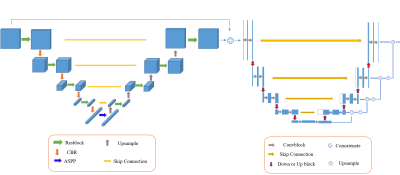
MRI study was performed on a 3T MR scanner (Discovery MR 750w, General Electric Healthcare, Waukesha, WI) using an 8-channel phase-array breast coil. All participants had undergone DCE-MRI using a three-dimension T1-weighted fast spoiled gradient echo sequence with the following imaging parameters: TR/ TE = 4.5/2.1 ms, field of view = 360 mm × 360 mm, image matrix = 512 × 512, slice thickness = 1.4 mm with no gap, flip angle = 12°. A total of 10 periods (35-55 s per period) with one pre-contrast and nine post-contrast dynamic periods were obtained. For DCE-MRI, gadoteric acid meglumine (Gd-DOTA, Dotarem, Guerbet, Roissy CdG Cedex, France) was injected at a rate of 2 mL/s (total dose, 0.1 mmol/kg of body weight) using a power injector (Ulrich, Germany) in the antecubital vein through a 20-gauge needle followed by a 20 mL saline flush. The structure of the proposed automated breast lesion segmentation framework was shown in Figure 1. A cascade framework was adopted, including 3D slice tough segmentation and 2D slice fine-tune segmentation. A 3D VNet and a 2D VNet was applied to breast lesion segmentation. At the same time, in order to prevent the influence of various sizes on the segmentation accuracy of the model, we also adopt a multi-scale dilation convolution module at the end of the encoding stage of 3D Vnet [5, 6]. In the training phase, firstly, the training data is input into 3D Vnet to obtain the rough segmentation results. In the second step, the training data and the rough segmentation results are cut into 2D slices in the transverse section according to the same cutting method, and the result cut by the input data are fused with the results cut by the rough segmentation results. The fused data is input into 2D Vnet, and the segmentation results obtained by 2D Vnet are spliced to obtain the final segmentation results. The loss function was defined as the sum of cross-entropy loss and dice loss. The performance of the proposed method for breast lesion segmentation was evaluated by comparing the automatically segmented results with the manual labels using dice similarity coefficient (DSC), and intersection-over-union (IoU).RESULTS AND DISCUSSION
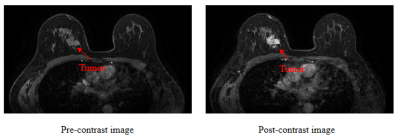
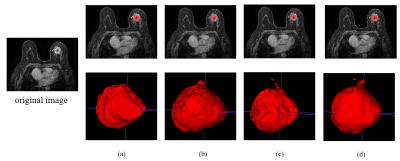
The results show that the proposed method can accurately and automatically segment the lesions on breast DCE-MRI with an average dice of 0.882 on the training set and 0.825 on the test set. As shown in Table 1, the average DSC and IOU of the proposed method on ten randomly selected test cases from the data set reaches 0.825 and 0.718, respectively. The values are higher than that of the Vnet, which achieves DSC and IOU of 803 and 0.677, respectively. The DCE images before and after contrast enhancement are shown in Figure 2. A representative segmentation result is shown in Figure 3. The second column is the segmentation result of VNet, and the segmentation result of the proposed cascade network is show in last column. The segmentation results of the proposed cascaded network are visually more similar to those manually delineated by doctors. The irregularity of the shape of breast lesions and the difference in size of different lesions make it difficult for the network to segment the lesions in DCE-MRI, especially the edges of the lesions. In this work, a cascade framework is proposed to automatically segment breast lesions from DCE-MRI. The cascade method of 3D convolution and 2D convolution is adopted to extract the spatial information in the MRI slice, thereby assisting the segmentation task of breast lesions. At the same time, we add dilated convolution with different dilation rate at the end of the encoding phase to extract the multi-scale information of the image, so as to solve the impact of tumor size differences on the segmentation results.Conclusion
The proposed network can realize automatic 3D segmentation of breast tumor and achieves a better segmentation result than traditional standard VNet, which provides an effective method for the accurate segmentation of breast lesions and hence the quantitative evaluation of follow-up clinical treatment effects.Acknowledgements
The study was partially support by National Natural Science Foundation of China (81830056), Key Laboratory for Magnetic Resonance and Multimodality Imaging of Guangdong Province (2020B1212060051), Guangdong Innovation Platform of Translational Research for Cerebrovascular Diseases, and Shenzhen Basic Research Program (JCYJ20180302145700745 and KCXFZ202002011010360).References
[1] Ferlay J, Colombet M, Soerjomataram I, et al. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods [J]. International journal of cancer, 2019, 144(8): 1941-1953.
[2] Pinker K, Helbich TH, Morris EA. The potential of multiparametric MRI of the breast. Br J Radiol 2017; 90: 20160715.
[3] Xiao J, Rahbar H, Hippe DS, Rendi MH, Parker EU, Shekar N, Hirano M, Cheung KJ, Partridge SC. Dynamic contrast-enhanced breast MRI features correlate with invasive breast cancer angiogenesis. NPJ Breast Cancer. 2021 Apr 16;7(1):42. doi: 10.1038/s41523-021-00247-3. PMID: 33863924; PMCID: PMC8052427.
[4] F Milletari, NavabN, Ahmadi S A. V-Net: Fully Convolutional Neural Networks for Volumetric Medical Image Segmentation[C]// 2016 Fourth International Conference on 3D Vision (3DV). IEEE, 2016.
[5] F Yu, Koltun V. Multi-Scale Context Aggregation by Dilated Convolutions[C]// ICLR. 2016.
Figures