3179
Deep Learning Algorithm for Automated Liver Segmentation Using Portal Venous Phase Magnetic Resonance Images1Department of Radiology, Beijing Friendship Hospital, Capital Medical University, Beijing, China, 2Shukun (Beijing) Technology Co., Ltd, Beijing, China
Synopsis
Accurate segmentation of liver not only facilitates the subsequent quantitative assessment of the regions of interest but also benefits precise diagnosis, and surgical planning. These tasks are usually performed by radiologists via visual inspection and manual delineations, which are tedious, labor-intensive, time-consuming. Convolutional neural networks (CNNs) have shown promise for performing automated liver segmentation for CT examinations, but there is less research on MR images. In this study, we provide a 3D U-Net based model for robust whole-liver and Couinaud segment measurements to support the treatment decision-making process on MR images.
Introduction
Liver segmentation from 3D abdominal scans is a crucial prerequisite for computer-aided interventions. Accurate segmentation not only facilitates the subsequent quantitative assessment of the regions of interest but also benefits precise diagnosis, prediction of prognosis, and surgical planning and intra-operative guidance[1]. These tasks are usually performed by radiologists via their visual inspection and manual delineations, which are tedious, labor-intensive, time-consuming, and prone to intra- and inter-observer variations. Therefore, automated segmentation algorithms are highly demanded to efficiently obtain accurate and reproducible segmentation results in routine clinical practice. Recently, convolutional neural networks (CNNs), a form of machine learning, have shown promise for performing automated liver segmentation for CT examinations[2,3]. However, there is less research on MR images. In this study, we would like to develop a 3D U-net based algorithm to automatically segment the liver into Couinaud regions on MR images.Methods
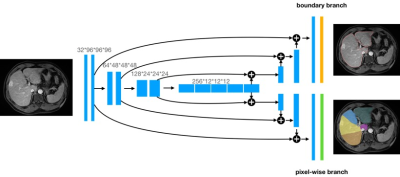
From 164 patients who underwent dynamic contrast-enhanced magnetic resonance 1.5T or 3.0T liver MRI examinations, 123 patients were randomly selected as the training dataset, and the other 41 patients as the test dataset to evaluate the performance of the final model. Classification of liver anatomy was based on Couinaud's description, which divides the organ into nine segments [4]. The ground-truth was generated by two radiologists with more than 5 years of experience in portal venous phase liver MR images by ITK-SNAP software. Our modified 3D U-net architecture for the liver segment segmentation is presented in Figure 1. 3D U-Net, which is built with 3D conv, ReLU, BN and pooling layers is used as the backbone. The output of the segmentation decoder has two channels. The upper segment produces the liver boundary and the other segment is the pixelwise segmentation of the liver segments. The model is trained end-to-end using the SGD optimizer on the training dataset. Dice loss function was used as the cost function. The segmentation accuracy of the liver was evaluated using the dice similarity coefficient (DSC) and volume ratio (RV).Results
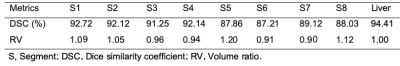
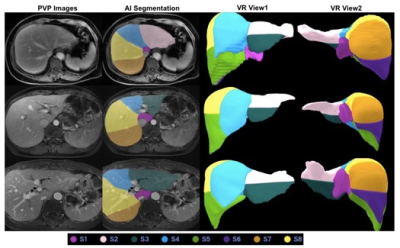
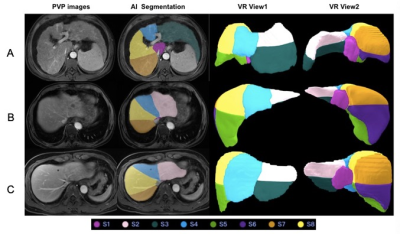
The dice loss and the segmentation accuracy of our model with liver and each segment are shown in Table 1. The accuracy for the DSC of different segment are between 87.21 and 92.72. The DSC of the whole liver segmentation is 94.41. And the volume ratio of the different segment ranged from 0.90 to 1.20. The representative results of automatic liver segmentation are presented in Figures 2,3. The average running time using the developed model on an GPU-accelerated computing platform with Xeon(R) Silver 4210R (CPU) and GeForce RTX 2070 (GPU) was 8.2 ± 2.4 s.Discussion
We present a novel, automated and efficient method for liver anatomy segmentation, which is a fundamental yet challenging problem in medical image analysis. This model generated good segmentation results a wide range of liver appearances from normal to abnormal. It is worth mentioning that some cases with advanced liver cirrhosis or multiple liver metastases were also accurately segmented by our model and some of the large across-segment tumors did not affect the detection of boundary lines. We recognize several limitations to this technical feasibility study. First, this study relied on retrospective data to train and validate the network. Thus, more research and validation may be required before a system like this can be deployed and relied on in clinical practice. Further work may be required to ensure similar performance across other scanner manufacturers, imaging phases, and institutional protocols. More extensive testing will help identify the failure modes of the 3D U-Net , thereby informing the selection of additional training data to further improve performance.Conclusion
Our study provides a 3D U-Net based model for robust whole-liver and Couinaud segment measurements to support the treatment decision-making process on MR images.Acknowledgements
NoReferences
[1] T. Heimann, B. Van Ginneken, M.A. Styner, et al. Comparison and evaluation of methods for liver segmentation from CT datasets IEEE Trans. Med. Imaging, 28 (8) (2009), pp. 1251-1265
[2] Kang Wang, Adrija Mamidipalli, Tara Retson, et al. Automated CT and MRI Liver Segmentation and Biometry Using a Generalized Convolutional Neural Network. Radiol Artif Intell. 2019 Mar; 1(2): 180022.
[3] Tian Y, Xue F, Lambo R, et al. Fully-automated functional region annotation of liver via a 2.5D class-aware deep neural network with spatial adaptation. Comput Methods Programs Biomed 2021;200:105818.
[4] T. Germain, S. Favelier, J.-P. Cercueil, et al. Liver segmentation: Practical tips. Diagnostic and Interventional Imaging, 2014,Volume 95, Issue 11,1003-1016.
Figures