3175
Synthetic-to-real domain adaptation with deep learning for fitting IVIM-DWI parameters1School of Medical Information Engineering, Guangzhou University of Chinese Medicine, Guangzhou, China, 2Department of Medical Imaging Center, Nanfang Hospital, Southern Medical University, Guangzhou, China
Synopsis
The intravoxel incoherent motion (IVIM) model of DWI with IVIM parameters has been widely used in characterization. However, the optimal method to obtain the IVIM parameters is still being explored. In this work, we propose a synthetic-to-real domain adaptation method for fitting the IVIM parameters. Specifically, we use synthesized data to train the network to learn the accurate mapping of the b-value images to the parameter map, and design a discriminator to help the network gradually adapt the learned mapping to the real data. Experimental results demonstrate that the proposed method outperforms previously reported methods for fitting IVIM parameters.
Introduction
Intravoxel incoherent motion(IVIM) has become widely used in the detection, characterization and staging of malignant lesions 1. The IVIM model uses a bi-exponential function to describe DW-MRI data, resulting a series of quantitative indicators including water molecule diffusion rate Dt, tissue perfusion (pseudo diffusion coefficient) Dp and tissue perfusion fraction Fp 2. Typically, the IVIM parameter map calculation is based on the voxel-by-voxel fitting the biexponential function, which is often prone to errors and calculation-intensive 3. Recently, supervised 4, unsupervised 5 and self-supervised 6 machine learning methods have been separately investigated for IVIM parameters estimation, but they still have their own shortcomings. For supervised learning, the synthesized data may have a large gap to the clinical data and the generalization of the network may be limited. For unsupervised learning and self-supervised learning methods, the accurate reconstruction of b-value images may not necessarily lead to the accurate estimation of IVIM parameters due to the motion and noise of IVIM-DWI. In this study, we propose a domain adaptation framework with convolutional neural network (CNN) for fitting the IVIM parameters.Material and Methods
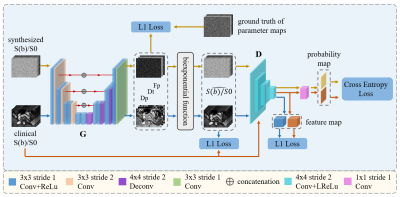
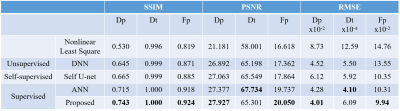
98 consecutive patients between January 2017 and September 2020 were included in the study, and routine IVIM-DWI serial examinations were performed using 3.0T MRI system (Achieva, Philips Healthcare, The Netherlands) in preoperative MR imaging. Axial DWI was performed with the following 9 b values (b = 0, 10, 20, 40, 80, 200, 400, 600, 1000s/mm2). As shown in Figure 1, the synthesized b-value images and the ground truth of their IVIM parameter maps are used to train the network to learn the accurate mapping from the b-value images to the IVIM parameter maps, and the clinical b-value images are used to assist the parameter map fitting network to gradually adapt the learned mapping to clinical data. Specifically, the network first fits the clinical b-value map to a parameter map, and then obtains the reconstructed b-value images through the bi-exponential equation 2, in which the loss function is designed optimize the quality of the reconstructed b-value images, thereby implicitly optimizing the quality of the IVIM parameter maps. In order to further adapt the synthetic data accurate mapping relationship to the clinical data, we designed a domain discriminator to distinguish between the clinical b-value images and the synthesized b-value images reconstructed by the parametric map fitting network. The parametric map fitting network is used as a generator to try to reconstruct the clinical b-value image that can deceive the discriminator. For the generation of synthesized data, we first uniformly sample the parameter map according to the following intervals: Dp between 0 and 0.2mm2/sec, Dt between 0.5×1e-3 and 2.5×1e-3 mm2/sec, and Fp between 0 and 90%. Then, we use the IVIM bi- exponential equation to get the normalized diffusion-weighted signal (S(b)/S0), and add complex Gaussian noise to simulate the Rician distribution. We use the Structural SIMilarity (SSIM) index and the Root Mean Square Error (RMSE) index to assess the similarity between the predicted map and the true value. In addition, the metric Peak Signal to Noise Ratio (PSNR) is used to assess the quality of the predicted maps. The proposed method is compared with the traditional nonlinear least square 3, ANN 4, unsupervised DNN 5 and Self-supervised U-net 6.Results
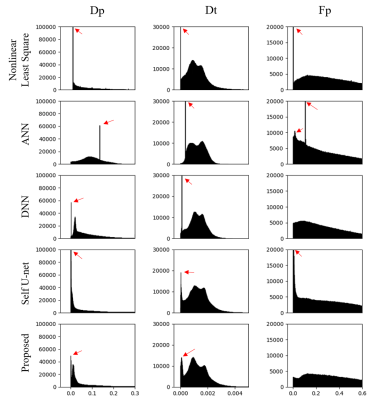
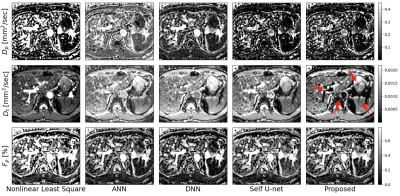
The experimental results are given in Table 1. First, ANN outperforms DNN, and Nonlinear Least Square (LS) yields the worst performance. In addition, self-supervised method obtained slightly better performance than the unsupervised method due to the utilization of CNN. It can be observed that the proposed method almost yield better performance than ANN, which attributes to the advantage of domain adaptation. Figure 2 shows the parameter distributions fitted on the clinical data of each method. It can be seen that the proposed method corresponds to the least outliers, which is more stable than other methods. Figure 3 shows the visualization of the parameter graphs fitted by different methods on clinical data. It can be seen that the proposed method fits more realistic textures.Discussion
The proposed method improves the fitting performance of IVIM parametric maps by making full use of the label information of synthesized data. Nonlinear Least Square method 3 yields the worst performance as it is prone to the motion and noise 1. Moreover, it is time consuming to fit the parameter map. Self-supervised method 6 obtained slightly better performance than the unsupervised method (DNN) 5 due to the utilization of spatial information with CNN rather than the voxel information in the DNN method. In addtion, self-supervision 6 is inferior to fully supervised learning ANN 4, indicating that purely self-supervised techniques is insufficient due to the motion and noise of IVIM-DWI. The proposed method almost yields better performance than ANN 4 due to the use of domain adaptation 7. Because the domain discriminator can effectively extract the key features of clinical data, so the parametric map fitting network can fit clearer and more meaningful texture information by optimizing the reconstruction loss function of feature map.Conclusion
In this study, we proposed a synthetic-to-real domain adaptation framework with deep learning for fitting the IVIM parameters, which can yield better performance than previously reported methods.Acknowledgements
This work is supported by the National Nature Science Foundation of China (81771920).References
1 Iima M, Bihan DL. Clinical intravoxel incoherent motion and diffusion MR imaging: past, present, and future. Radiology 2016; 278(1):13-32.
2 Bihan DL, Breton E, Lallemand D, Aubin ML, Vignaud J, Laval-Jeantet M. Separation of diffusion and perfusion in intravoxel incoherent motion MR imaging. Radiology 1988; 168(2):497-505.
3 Lanzarone E, Mastropietro A, Scalco E, Vidiri A, Rizzo G. A novel bayesian approach with conditional autoregressive specification for intravoxel incoherent motion diffusion-weighted MRI. NMR in Biomedicine 2020;33:e4201.
4 Bertleff M, Domsch S, Weingartner S, et al. Diffusion parameter mapping with the combined intravoxel incoherent motion and kurtosis model using artificial neural networks at 3T. NMR in Biomedicine 2017; 30(12):e3833.
5 Barbieri S, Gurney-Champion OJ, Klaassen R, Thoeny HC. Deep learning how to fit an intravoxel incoherent motion model to diffusion-weighted MRI. Magnetic resonance in medicine 2020;83(1): 312 -321.
6 Vasylechko SD, Warfield SK, Afacan O, Kurugol S. Self-supervised IVIM DWI parameter estimation with a physics based forward model. Magnetic resonance in medicine 2021 (October 22 online)
7 Bousmalis K, Silberman N, Dohan D, et al. Unsupervised Pixel-Level Domain Adaptation with Generative Adversarial Networks. IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 2017: 95-104.
Figures