3159
A unified deep learning method for 4D-MRI motion deformation estimation and image enhancement1Department of Health Technology and Informatics, Hong Kong, China
Synopsis
We proposed a unified deep learning framework for predicting the motion deformation between different phases of 4D-MRI with simultaneous image quality enhancement. The network combines a coarse-to-fine unsupervised registration model to estimate the deformation vector fields (DVFs) in different image scales and a GAN-based enhancement network to restore anatomic features. Particularly, a prior knowledge of 4D-MRI is incorporated into the unified model, guiding an accurate DVF prediction and maintaining image topology. Both qualitative and quantitative results showed that the predicted DVFs and resultant 4D-MRI images achieved improved performance compared with the traditional method without modifications.
INTRODUCTION
Four-dimensional magnetic resonance imaging (4D-MRI) technique has been highly desirable in the radiotherapy workflow for its excellent soft-tissue contrast. Especially, 4D-MRI image-guided radiation therapy (IGRT) is preferred for treating tumors affected by respiratory motion1,2. As one of the promising imaging techniques in 4D-MRI, prospective 4D-MRI achieves fast volumetric acquisition. However, for the sake of rapid imaging speed, such 4D-MRI provides limited spatial resolution and a relatively low signal-to-noise ratio, negatively affecting the estimation accuracy of deformable vector fields (DVF) between different phases/frames and further hindering the application of 4D-MRI in IGRT3. To overcome the aforementioned deficiencies, this method proposes a unified end-to-end network, which aims to predict DVFs among any phases in the 4D-MRI sequence while boosting MRI quality simultaneously. For the spatial resolution, the proposed network can recover more detailed spatial information with the help of a normal MRI of the same patient with high resolution. Moreover, the generated 4D-MRI sequence enables accurate temporal resolution, which benefits from the proposed prior image-guided coarse-to-fine model in an unsupervised manner.METHODS
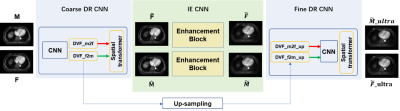
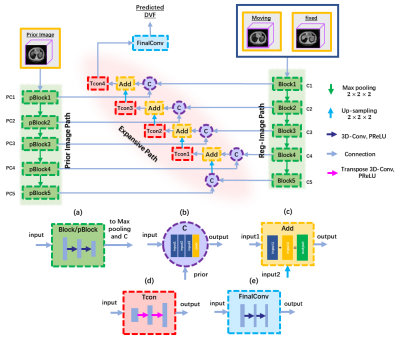
Figure 1 illustrates the design of the framework, which consists of three modules, including a coarse DR CNN, an image enhancement (IE) CNN, and a fine DR CNN. The integration of a coarse CNN and a fine DR CNN perform an unsupervised coarse-to-fine registration strategy to predict the DVF between any two phases of images in 4D-MRI sequence. Before updating DVFs in the fine DR CNN, an IE CNN is adopted to restore finer details of the misaligned image pairs.The coarse DR CNN inputs any two phases of images in 4D-MRI, denoted as the moving images (M) and fixed images (F), and tries to predict the DVF $\emptyset$ and $\emptyset^{T}$ between them in bi-direction. The network was trained in an unsupervised manner4, utilizing the VoxelMorph5 network architecture. The loss function measures image similarity between the deformed image $M\circ\emptyset$ or $F\circ\emptyset^{T}$ and its corresponding target image $F$ or $M$. In IE CNN, we adopted a classical pixel2pixl network6 to enhance the image quality of 4D-MRI. The network inputs 2D high-quality and low-quality image pairs for training without considering the temporal correlation among each phase in 4D-MRI. The reason for using 2D image pairs in the transversal view is that limited MRI volumes were obtained.Finally, the predicted coarse DVFs and the enhanced MRI image pairs were together fed into the fine DR CNN. Unlike the coarse DR CNN, we added an independent feature extraction channel by incorporating a normal MRI image called prior MRI, which is commonly available when conducting an MRI scan of the same patient. Note that the prior MRI and enhanced fixed image should be the same frame/phase, so a pre-registration process based on ELASTIX toolbox was needed7. Figure 2 illustrates the detailed information of the fine DR CNN, which is also called N-Net developed in our previous study8. By doing so, the fine DR CNN obtained a finer DVF estimation in cooperation with the prior MRI image with tiny anatomic features.
The dataset used for training and testing the proposed network were obtained from twenty patients undergoing radiotherapy for liver tumors. The study protocol was approved by the institutional review board. Each patient underwent 4D-MRI using the TWIST volumetric interpolated breath-hold examination (TWIST-VIBE) MRI sequence. Then ten amplitude-correlated phases were selected from the original 4D-MRI sequence by the body area method9. All the patients were also underwent a regular T1w 3D MRI scan with highly spatial image resolution and were regarded as the prior image of each patient. Of the twenty patients, eighteen cases were used as training, while the others were used for testing.
RESULTS
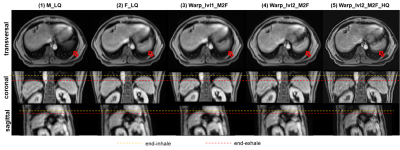
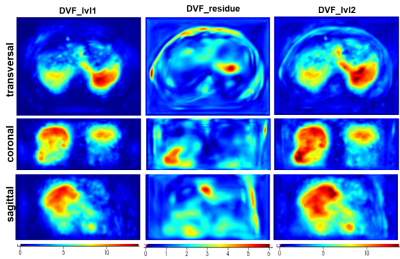
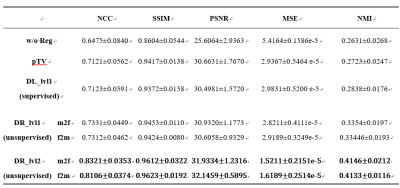
Figure 3 displays the intermediate results of the proposed model in three different views. As shown in Fig.3(4), an updated deformed image with a finer DVF was obtained compared to the warped image by the coarse DR CNN in Fig.3(3). Moreover, a refined prediction of the warped image from the opposite direction also achieved satisfactory alignment. In addition, both the results in Fig.3(4) and Fig.3 (5) recovered more detailed anatomic features in contrast to the corresponding coarse DR-based images, indicating the feasibility of the proposed N-Net. Figure 4 shows the heatmap of the absolute magnitude value of coarse DVF, updated residual DVF, and the final refined DVF. It can be observed that the fine DR CNN could upgrade the estimation of DVF with more detailed deformations at large displacement positions. Detailed quantitative evaluation results are listed in Table 1, indicating that the proposed coarse DR CNN has achieved better performance than the traditional supervised VoxelMorph method. Furthermore, by adding the fine DR CNN, the final registration results showed improved image similarity with the fixed image in terms of all the evaluation metrics, compared with pTV10, supervised VoxelMorph, and coarse DR CNN.DISCUSSION&CONCLUSION
For accurately estimating motion deformation and simultaneously improving the image quality of 4D-MRI, we developed and evaluated a unified deep learning framework that integrates a coarse-to-fine DR CNN and an EI CNN. Experimental results demonstrated that our model achieved satisfactory performance in both DVF prediction and image quality enhancement for 4D-MRI at the same time.Acknowledgements
No acknowledgement found.References
1. Cai J, Chang Z, Wang Z, Paul Segars W, Yin FF. Four-dimensional magnetic resonance imaging (4D-MRI) using image-based respiratory surrogate: A feasibility study. Med Phys. 2011;38(12):6384-6394. doi:10/fkx5pm
2. Wang C, Yin FF. 4D-MRI in Radiotherapy. In: Manchev L, ed. Magnetic Resonance Imaging. IntechOpen; 2019. doi:10.5772/intechopen.84592
3. Stemkens B, Paulson ES, Tijssen RHN. Nuts and bolts of 4D-MRI for radiotherapy. Phys Med Biol. 2018;63(21):21TR01. doi:10/gjm6vk
4. Balakrishnan G, Zhao A, Sabuncu MR, Guttag J, Dalca AV. An Unsupervised Learning Model for Deformable Medical Image Registration. 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition. Published online June 2018:9252-9260. doi:10/ggchmh
5. Balakrishnan G, Zhao A, Sabuncu MR, Guttag J, Dalca AV. VoxelMorph: A Learning Framework for Deformable Medical Image Registration. IEEE Transactions on Medical Imaging. 2019;38(8):1788-1800. doi:10/gfx7zs
6. Isola P, Zhu JY, Zhou T, Efros AA. Image-to-Image Translation with Conditional Adversarial Networks. arXiv:161107004 [cs]. Published online November 26, 2018. Accessed August 3, 2021. http://arxiv.org/abs/1611.07004
7. Klein S, Staring M, Murphy K, Viergever MA, Pluim JPW. elastix: A Toolbox for Intensity-Based Medical Image Registration. IEEE Transactions on Medical Imaging. 2010;29(1):196-205. doi:10/ds6xss
8. Zhi S, Kachelrieß M, Pan F, Mou X. CycN-Net: A Convolutional Neural Network Specialized for 4D CBCT Images Refinement. IEEE Transactions on Medical Imaging. 2021;40(11):3054-3064. doi:10/gkmssm
9. Liu Y, Yin FF, Chang Z, et al. Investigation of sagittal image acquisition for 4D-MRI with body area as respiratory surrogate. Medical Physics. 2014;41(10):101902. doi:10/gjp27v
10. Vishnevskiy V, Gass T, Szekely G, Tanner C, Goksel O. Isotropic Total Variation Regularization of Displacements in Parametric Image Registration. IEEE Transactions on Medical Imaging. 2017;36(2):385-395. doi:10/gjww6z
Figures