3157
Developing a comprehensive whole body MR protocol and model generation pipeline for children1Mātai Medical Research Institute, Gisborne, New Zealand, 2General Electric Healthcare AUS/NZ, Melbourne, Australia, 3Auckland Bioengineering Institute, Auckland, New Zealand, 4Faculty of Medical and Health Sciences, University of Auckland, Auckland, New Zealand, 5Ngāi Tāmanuhiri, Tairāwhiti, New Zealand, 6Rongowhakaata, Tairāwhiti, New Zealand, 7Ngāti Pāhauwera, Hawke's Bay, New Zealand, 8University of Otago, Dunedin, New Zealand, 9Ngāi Tahu, Te Waipounamu, New Zealand, 10Kāti Māmoe, North Otago, New Zealand, 11Mātai Ngā Māngai Māori, Gisborne, New Zealand, 12Te Aitanga a Hauiti, Tairāwhiti, New Zealand, 13Ngāti Apa, Rangitīkei, New Zealand, 14Massey University, Palmerston North, New Zealand, 15Ngāti Kahungunu, Wairarapa, New Zealand, 16Rongomaiwahine, Mahia, New Zealand, 17Ngāti Porou, Tairāwhiti, New Zealand, 18Ngāti Kahungunu, Wairoa, New Zealand, 19University of Cantebury, Christchurch, New Zealand, 20Ngāpuhi, Te Tai Tokerau, New Zealand, 21Turanga Health, Tairāwhiti, New Zealand
Synopsis
Approaches to magnetic resonance (MR) image acquisition and computational modelling often exist in silo. The application of computational modelling to paediatric data is in its infancy. This collaborative study addresses this gap for multiple organ systems. State-of-the art MR imaging sequences were selected and refined to provide imaging data for the creation of subject-specific geometric models. The MR protocol and modelling outputs will underpin a longitudinal paediatric study set in Aotearoa New Zealand, generating normative data in healthy children, and identifying early biomarkers of pathological processes. This will enable the development of new personalised, predictive, and preventative models of care.
INTRODUCTION
Understandings of child development are limited by a lack of comprehensive normative imaging.1,2,3 MR imaging is the superior imaging modality for children, as it is non-ionising. However, paediatric cohorts are often difficult to scan due to motion and reduced adherence to breathing control requests during scanning. Longitudinal paediatric magnetic resonance imaging (MR) studies have predominantly focused on neuroimaging, with few studies imaging across multiple organs and multiple time points.5Computational modelling greatly enhances the value of imaging outputs, improving their research and clinical utility.6 High quality, fast multi-organ MR imaging, coupled with personalized computational modeling could deliver important clinical applications. So far, most imaging and modelling research has existed in silo, developed from clinical needs to study individual organs or systems. However, to address complex physiological questions, multiscale models of multiple organ systems are needed.7 Such personalized models that encompass inter-organ relationships are currently being developed using next-generational biophysical modelling frameworks.8
In this study, we developed and tested a whole-body imaging protocol to scan young children aged 8-11. Initial computational modelling outputs were developed in tandem, with iterative feedback supporting protocol development.
METHODS
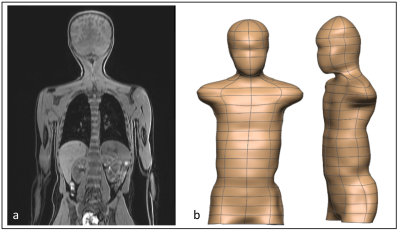
We have developed a comprehensive MRI imaging protocol for brain, cardiovascular, vascular, abdominal, and musculoskeletal organ systems and employed state-of-the-art processing pipelines for generating computational models. MR images were acquired in a female child (Age=8yr; Weight=29Kg) using a 3T GE Premier scanner (General Electric, MI, USA) using state-of-the-art 48-channel head coil and Air Coil (AIR™) technologies based at Mātai, Gisborne-Tairawhiti, New Zealand. In-bore screen, acoustic shielding, and pre-scan familarisation were used to improve children’s experience. approval was granted through the Health and Disability Ethics Committee (20/CEN/107). Local approval has been granted via the Mātai Ngā Māngai Māori Advisory Board. Imaging sequences were selected and iteratively refined to achieve short acquisition times. For every sequence, test-retest was carried out in 2 children (results not reported) to check for consistent measurements and to ensure adequate image quality and reproducibility.RESULTS
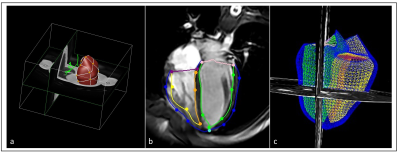
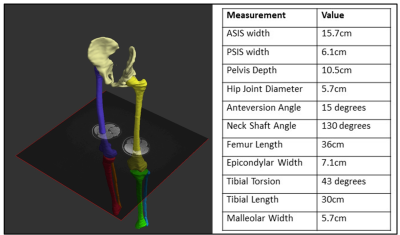
Following test-retest and refinement of images and model generation, a total of 20 sequences were finalized to cover whole body. Table 1 lists parameters from the core modelling sequences. The remaining sequences formed part of the standard acquisition, which will either be incorporated into the modelling pipelines or used in other data pathways. Figures 1-4 show the modelling outputs derived from the MRI imaging, ranging from 3D brain volumes and cardiac models to lung and musculoskeletal models. Models of the paediatric neurovasculature are also displayed.DISCUSSION
The MRI protocol and modelling have been designed to achieve scalability towards a longitudinal study through use of state-of-the-art 3T MRI technology.The application of computational modelling to paediatric MRI data is in its infancy.6 This preliminary study demonstrates the use of multiple imaging sequences across key organ systems with results from associated computational models. A robust workflow has been developed through this study, with the creation of personalised 3D models that allow for in-depth analysis.
While a key part of the value and innovation of the study lies in affording analysis across organ systems, examples of the individual value of modelling for each system is outlined below:9
Brain: Contributing to a definitive set of longitudinal percentile normative curves for brain growth across a wide range of structures. Accurate, automated reporting of brain volumes promises to help understand the impact of childhood environments on brain development.
Cardiac: Personalized biventricular models allow for more in-depth analysis of cardiac function. Standard measurements include mass, volume, and strain. However, models of shape and motion can be used to create population based normative models against which individuals can be compared, and further biomarkers derived.
Lung: Computed tomography uses ionizing radiation, and as such is not typically suited for paediatric research. Ultra-short echo imaging has been shown to provide useful structural information of the lung. Imaging during free-breathing provides information on thoracic cavity shape and volume change.
Abdomen: Automated segmentation of abdominal viscera and fat is in its infancy; more so for the application of these techniques to paediatric data. This will improve our understanding of individual differences in fat distribution and relationships with metabolic health.
Musculoskeletal: Subject-specific modelling of bone and muscle growth will allow a better understanding of typical musculoskeletal development in children, helping to answer key questions such as, ‘how do bone measurements evolve through growth?’.
Vascular: Whole-body vascular imaging and deep learning-based automatic segmentation of the vasculature has been validated and is ready for application to larger scale studies. These will enable an understanding of relationships between vascular and organ growth and development, and ultimately shed light on pathological processes.
CONCLUSION
This study is the first of its kind applying a modelling approach and advanced imaging to multiple organs in children. Embedding computational modelling into the design of a future longitudinal imaging study will build on the success of previous longitudinal studies.The technical and operational experience gained form this process has resulted in the development of an MR imaging protocol ready for deployment in a pioneering cohort study. The imaging data to model pipeline will allow for an in-depth probing and predictive analysis of structure-function relationship in children of the Tairāwhiti-Gisborne region of New Zealand.
Acknowledgements
Kānoa - Regional Economic Development & Investment Unit, New Zealand
Trust Tairāwhiti, Gisborne, New Zealand
Advice and support from GE Healthcare is acknowledged
*Denotes equal authorship contributions
References
1. Tamnes CK, Roalf DR, Goddings AL, et al. Diffusion MRI of white matter microstructure development in childhood and adolescence: Methods, challenges and progress. Developmental Cognitive Neuroscience. 2018;33:161–75.
2. Fernandez J, Dickinson A, Hunter P. Population based approaches to computational musculoskeletal modelling. Biomechanics and Modeling in Mechanobiology. 2020;19:1165–8.
3. van der Ven JPG, Sadighy Z, Valsangiacomo Buechel ER, et al. Multicentre reference values for cardiac magnetic resonance imaging derived ventricular size and function for children aged 0-18 years. European Heart Journal Cardiovascular Imaging. 2020;21:102–13.
4. Wood-Frohlich L, Martin T, Malisza K. Training Children to Reduce Motion and Increase Success of MRI Scanning. Current Medical Imaging Reviews. 2010;6:165-170.
5. White T, Muetzel RL, el Marroun H, et al. Paediatric population neuroimaging and the Generation R Study: the second wave. European Journal of Epidemiology. 2018;33:99–125.
6. Cerrolaza JJ, Picazo ML, Humbert L, et al. Computational anatomy for multi-organ analysis in medical imaging: A review. Medical Image Analysis. 2019;56:44–67.
7. P. Hunter, "The Virtual Physiological Human: The Physiome Project Aims to Develop Reproducible, Multiscale Models for Clinical Practice. IEEE Pulse. 2016;7:4;36-42.
8. B. G. Beutel, S. J. Girdler, J. A. Collins, N. Y. Otsuka, and A. Chu. Characterization of proximal femoral anatomy in the skeletally-immature patient. Journal of Child Orthopaedics. 2018;12:167–172.
9. Osanlouy, M., Bandrowski, A., De Bono, B., Brooks, D., Cassarà, A. M., Christie, R., ... & Hunter, P. J. The SPARC DRC: Building a Resource for the Autonomic Nervous System Community. Frontiers in Physiology. 2021;12:693-735.
Figures
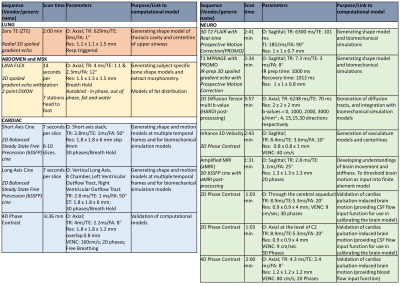
Table 1: MRI Protocol. Key sequences used to underpin modelling outputs are detailed here, with scan time, salient imaging parameters and links to computational models stated.
O: Orientation; TR: Time repetition; TE: Time echo; TI: Inversion time interval; FA: Flip angle; Res: resolution;VENC: Velocity encoding
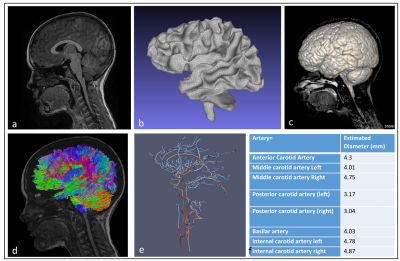
Figure 1: (a) Mid-sagittal T1 MP-RAGE sequence (b) White matter tissue segmented using FSL based on 3D Cube T2 FLAIR sequence (c) 3D brain parenchyma volume rendering overlaid on mid-sagittal slice from T2 FLAIR. (d) Probabilistic tractography generated using MRtrix overlaid on mid-sagittal T1 MP-RAGE. (e) subject-specific skeletonisation of cerebral arterial tree (3D Axial Inhance Velocity) using in-house code obtaining geometrical network information (diameter colour-coded). (f) displays vascular metrics extracted from computational model of the cerebral arterial tree.