3102
Predicting Knee Osteoarthritis Progression with an Interpretable Deep Learning Approach on MRIs: Data from the Osteoarthritis Initiative1Department of Medical Imaging, The Third Affiliated Hospital of Southern Medical University, GuangZhou, China, 2Department of Computer Science & Engineering, The Chinese University of Hong Kong, HongKong, China, 3Philips Healthcare, GuangZhou, China
Synopsis
Identifying knee osteoarthritis progressors is significant. MRIs can reflect the structures of the knee. However, currently no tool could rapidly and objectively predict knee osteoarthritis progression based on MRI. Therefore, we applied deep learning algorithms on MRIs of the whole knee to predict progression at three time points. The Gradient-weighted Class Activation Maps were employed for interpretability, and the highlighted infrapatellar fat pad (IPFP) was segmented for progression prediction. We showed that the deep learning framework performed well on discrimination of progressors, especially at 24th months, and that the infrapatellar fat pad plays an important role in predicting progression.
abstract
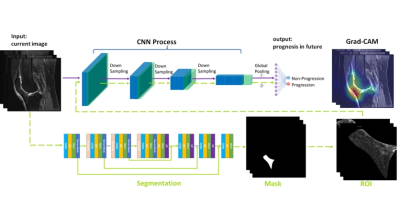
Purpose and Introduction Knee osteoarthritis (KOA) is one of the most common diseases among the elderly. However, there are no licensed disease-modifying drugs for treatment[1]. Progressors, who can better reflect therapy efficacy, are the best candidates for novel disease-modifying trials[2]. Also, identifying high-risk progressors and providing initiative physical therapies at an early stage is extremely beneficial[3, 4]. Therefore, it is critical to detect the progressors with a high risk of structural and pain progression. MRI, which can reveal changes of sophisticated architectures of the knee, may be used as a tool for detecting those who will progress. The current studies predicting KOA progression are either based on semi-quantitative grading or quantitative measuring of MRI structures. However, such methods are difficult to broaden to a larger population because of the heavy workload. Deep learning algorithms may be employed as a feasible option for this challenge. Therefore, our study aims to develop a deep learning based predictive tool for KOA progression and further expose structures responsible for the prediction.Materials and MethodsMRIs of 364 knees (mean age, 62 years ± 9) were obtained from the Osteoarthritis Initiative and retrospectively included in this study, with 182 knees progressing both in radiology and symptoms, and 182 knees neither not. The criteria of radiographic progression means that the medial joint space width loss less than 0.7mm during the first 24-months but more than 0.7mm over the last 24-months. For pain progression, it satisfied the criterion that the knee pain subscale assessed by the Western Ontario and McMaster Universities Osteoarthritis Index questionnaire did not increase above a minimum clinically important difference (9 points or more) during the first 24-months but did increase over the last 24-months[5-7]. Three well-known networks: 3D Resnet50, 3D Resnet (2+1) d50, and 3D DenseNet169 [8, 9] were applied on the same prediction task at three time points. All the deep learning models are trained only with patient-level labels without labeling the specific lesion locations and signal abnormalities in an image. The 5-fold cross-validation was used for all classifiers to bias and generalize the results, we split data into two parts, leaving aside about 20% of the data for model evaluation and 80% of the data to select the optimal model and we repeated this procedure 5 times until all the data was used for testing. The details of our image-alone models and complex models are shown in Figure 1. Furthermore, to explain the regions that are important for the CNN algorithm’s decisions, the Gradient-weighted Class Activation Maps (Grad- CAM) were used. As the IPFP was highlighted by the Grad-CAM and its segmentation is not the same as the other regions such as patellofemoral or tibiofemoral joints that require dividing into subregions, the infrapatellar fat pad was selected to explore its function for predicting KOA progression and compared with the whole knee. We evaluate our models using widely-acknowledged metrics: (1) Area under the Curve of ROC (AUC), (2) Sensitivity, (3) Specificity. Wilcoxon Signed Ranks Tests[8] were used for comparing parameters between two pair classification models, whereas Friedman Tests were employed to evaluate longitudinally paired models.
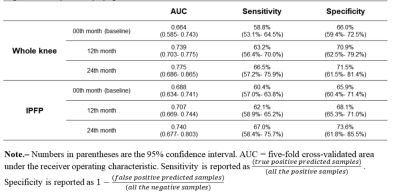
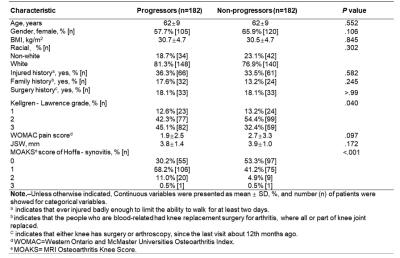
Results The demographics of the participants are shown in Table 1. The results of the test data for three architectures are summarized in Table 2. 3D Resnet 50, 3D Resnet (2+1) d50, and 3D DenseNet 169 achieved AUCs with 0.780, 0.786, and 0.775 at 24th month, 0.738, 0.725, and 0.739 at 12th month, and 0.640, 0.653, and 0.664 at baseline, respectively. The CNN with infrapatellar fat pad predicted KOA progression with 0.740, 0.707, 0.688 at 24th month, 12th month, and baseline respectively. The comparison of the infrapatellar fat pad and the whole knee is detailed in Table 3.
Conclusion From the whole knee MRIs, we confirmed the possibility of employing deep learning networks for predicting knee osteoarthritis progression and made it interpretable. Furthermore, through developing predictive models based on the infrapatellar fat pad, the function of the infrapatellar fat pad in predicting knee osteoarthritis was verified.
Acknowledgements
Jiaping Hu and Chuanyang Zheng contributed equally to this work.
Xiaodong Zhang and Qi Dou are both co-corresponding authors.
References
1. Roemer FW, Collins J, Kwoh CK, Hannon MJ, Neogi T, Felson DT, et al. MRI-based screening for structural definition of eligibility in clinical DMOAD trials: Rapid OsteoArthritis MRI Eligibility Score (ROAMES). Osteoarthritis Cartilage 2020, 28(1): 71-81.
2. Widera P, Welsing PMJ, Ladel C, Loughlin J, Lafeber F, Petit Dop F, et al. Multi-classifier prediction of knee osteoarthritis progression from incomplete imbalanced longitudinal data. Sci Rep 2020, 10(1): 8427.
3. Guan B, Liu F, Haj-Mirzaian A, Demehri S, Samsonov A, Neogi T, et al. Deep learning risk assessment models for predicting progression of radiographic medial joint space loss over a 48-MONTH follow-up period. Osteoarthritis Cartilage 2020, 28(4): 428-437.
4. Mobasheri A, Rayman MP, Gualillo O, Sellam J, van der Kraan P, Fearon U. The role of metabolism in the pathogenesis of osteoarthritis. Nat Rev Rheumatol 2017, 13(5): 302-311.
5. Hunter DJ, Nevitt M, Losina E, Kraus V. Biomarkers for osteoarthritis: current position and steps towards further validation. Best Pract Res Clin Rheumatol 2014, 28(1): 61-71.
6. Eckstein F, Collins JE, Nevitt MC, Lynch JA, Kraus VB, Katz JN, et al. Brief Report: Cartilage Thickness Change as an Imaging Biomarker of Knee Osteoarthritis Progression: Data From the Foundation for the National Institutes of Health Osteoarthritis Biomarkers Consortium. Arthritis Rheumatol 2015, 67(12): 3184-3189.
7. Angst F, Aeschlimann A, Stucki G. Smallest detectable and minimal clinically important differences of rehabilitation intervention with their implications for required sample sizes using WOMAC and SF-36 quality of life measurement instruments in patients with osteoarthritis of the lower extremities. Arthritis Rheum 2001, 45(4): 384-391.
8. Hara K, Kataoka H, Satoh Y. Can spatiotemporal 3d cnns retrace the history of 2d cnns and imagenet? Proceedings of the IEEE conference on Computer Vision and Pattern Recognition; 2018; 2018. p. 6546-6555.
9. Tran D, Wang H, Torresani L, Ray J, LeCun Y, Paluri M. A closer look at spatiotemporal convolutions for action recognition. Proceedings of the IEEE conference on Computer Vision and Pattern Recognition; 2018; 2018. p. 6450-6459.
10. Choi KS, You SH, Han Y, Ye JC, Jeong B, Choi SH. Improving the Reliability of Pharmacokinetic Parameters at Dynamic Contrast-enhanced MRI in Astrocytomas: A Deep Learning Approach. Radiology 2020, 297(1): 178-188.
Figures
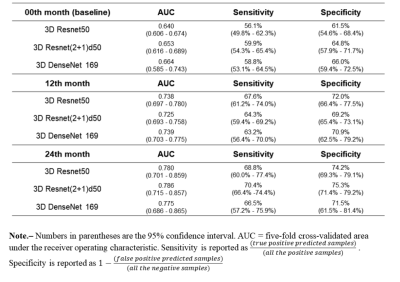
Results of three architectures performed in the whole knee MRIfor predicting progression at three time points