3097
Clinical Feasibility Study of Accelerated 2D Magnetic Resonance shoulder Imaging Using Deep Learning-based Algorithm
Jing Liu1, Ke Xue2, Yongming Dai2, Peng Wu2, and Jianxing Qiu1
1Peking University First Hospital, Beijing, China, 2MR Collaboration, Central Research Institute,United Imaging Healthcare, Shanghai, China
1Peking University First Hospital, Beijing, China, 2MR Collaboration, Central Research Institute,United Imaging Healthcare, Shanghai, China
Synopsis
Deep Learning-based magnetic resonance imaging (DL-MRI) could accelerate 3D MRI scanning time. In this study, we investigate the feasibility of DL-MRI in 2D shoulder MRI. Totally 20 consecutive patients were enrolled for both conventional MRI and DL-MRI. Both qualitative and quantitative analyses were conducted to compare the image quality and lesion diagnosis on conventional MRI and DL-MRI. And our results revealed that DL-MRI was valuable for improving the overall workflow of shoulder MRI with scanning time saved and image quality improved.
Introduction
To evaluate the clinical feasibility of 2D shoulder magnetic resonance imaging (MRI) using deep learning (DL) algorithm in comparison with conventional MRI (Con-MRI).Methods
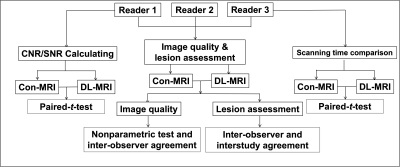
This prospective study was approved by local ethics committee. From August 2021 to November 2021, 20 consecutive patients (12 males, 8 females, mean age: 52±18 years, age range: 30-81 years) with suspected disease on shoulder were enrolled. The MRI examinations included both Con-MRI (Sagittal T2 weighted imaging (T2WI), Axial T1WI and proton density weighted imaging (PDWI), Coronal T1WI and PDWI) and DL-MRI (with 2.5 factor for acceleration: Sagittal T2 weighted imaging (T2WI), Axial T1WI and PDWI, Coronal T1WI and PDWI) for each patient. All MR images were acquired with a 3T MR scanner (uMR790, United Imaging Healthcare, Shanghai, China ). The neural network used in this study was trained based on the designed ResNet1 with millions of under-sampled data as input and corresponding fully-sampled data as learning target. Detail information of this network was described in a previous study2.Three radiologists (21, 13 and 7 years of experience in skeletal radiology, respectively) independently assessed the image quality with blind to the information of patients’ data. Image quality was rated according to a 5-point Likert scale:1) Non-diagnostic image quality due to severe artifacts, image distortion, or poor signal intensity.2) Poor diagnostic image quality and diagnostic confidence of the readers due to severe artifacts, image distortion pronounced, or relatively poor signal intensity.3) Moderate image quality and confidence of the readers due to moderate artifacts, moderate image distortion.4) Good diagnostic image quality: few artifacts, slight image distortion, the main structures were well displayed.5) Excellent image quality: almost no artifacts or imaging distortion; the main structures were perfectly displayed.Both signal-to-noise ratio (SNR) and contrast-to-noise ratio (CNR) for bone marrow and surrounding soft tissue were assessed for both DL-MRI and Con-MRI. Signal intensity (SI) was measured as following: round-shaped regions of interest (ROIs) were placed on the humeral head and surrounding muscle. Another round-shaped ROI was placed on the air far away from the shoulder, to calculate the standard deviation (SD) of air as background noise.Three readers assessed them separately in a blinded and randomized fashion. The interobserver and interstudy agreement were assessed for evaluation of lesions with reader 1 as reference, who has more diagnostic experience. The flowchart is shown in Figure 1.Statistical analysis was performed by using statistical software (SPSS, v. 24.0; Chicago, IL). A Kendall W test was performed to assess the interobserver agreement of scores of image quality and lesion diagnosis between readers. A nonparametric test (Wilcoxon test) was performed to compare the image quality between Con-MRI and DL-MRI. For lesion diagnosis, the interobserver agreement betweenreaders and interstudy agreement between Con-MRI and DL-MRI for each observer were evaluated by kappa analysis. Paired t-tests for SNR and CNR calculation were conducted to compare Con- MRI and DL- MRI.Results
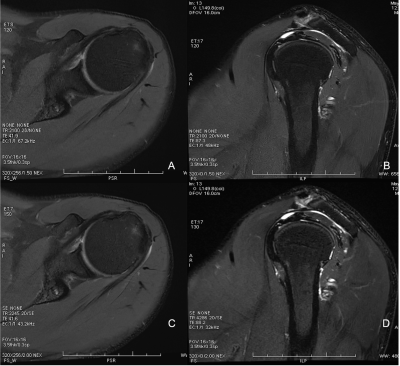
We had achieved totally 11min25sec and 6min1sec by using Con and DL protocols (t = -47.464, P < 0.0001), respectively. Excellent Interobserver agreement was found for image quality on CS-MRI (Kendall W value of 0.879, P < 0.001) and Con-MRI (Kendall W value of 0.926, P < 0.001). The image quality showed no statistical significance between DL-MRI and Con-MRI (the mean scores for DL-MRI of readers 1, 2, and 3 were 4.82±0.44, 4.86 ±0.35, and 4.78±0.47, respectively; for Con-MRI were 4.81 ± 0.53, 4.87 ± 0.58, and 4.76 ± 0.62, respectively) (Z = 3.710, with P = 0.58). Table 1 shows the number of lesions which were diagnosed by readers on the Con-MRI and DL-MRI. There was excellent interobserver agreement in the diagnosis for Con-MRI (Kendall W value of 0.903, P < 0.001) and DL-MRI (Kendall W value of 0.916, P < 0.001). Figures 2 display the examples of clear imaging both on Con-MRI and DL-MRI.The paired t-test showed significant variance results between Con-MRI and DL-MRI on each sequence. The SNRs of bone marrow comparison were: (413.78 ± 70.17) of Con-MRI and (443.71 ± 71.9) of DL-MRI (t = –5.264, P < 0.001). The CNRs of bone marrow comparison were: (152.54 ± 54.16) of Con-MRI and (178.64 ± 59.59) of DL-MRI (t = –5.544, P < 0.001).Discussion
As a popular MRI technique with reduced scan time, CS has been widely used. However, most of previous studies emphasized on its feasibility in 3D imaging or single MR sequence. Due to the principle of sparse under-sampling, CS technique was reasonable in improving the work efficacy of 3D MRI. However, 3D MRI was not applicable to all clinical setting, such as brain, spine, and abdominal imaging. In some applications, 2D MRI is more frequently used. Therefore, reducing the scan time with comparable or better image quality in 2D MRI needs further investigation.Conclusion
We demonstrated that compared with Con-MRI, DL-MRI could save scan time significantly without loss of image quality in the routine clinical shoulder examinations. In conclusion, our results suggested that DLMRI was valuable for improving the overall workflow of shoulder MRI.Acknowledgements
N/AReferences
[1] He K, Zhang X, Ren S, et al. Deep residual learning for image recognition. Proceedings of the IEEE conference on computer vision and pattern recognition. 2016: 770-778.[2] Sheng R, Zheng L, Jin K, et al. Single-breath-hold T2WI liver MRI with deep learning-based reconstruction: A clinical feasibility study in comparison to conventional multi-breath-hold T2WI liver MRI. Magnetic Resonance Imaging, 2021.
DOI: https://doi.org/10.58530/2022/3097