3062
Radiomics analysis of neurite orientation dispersion and density imaging: differentiation between glioblastoma and solitary brain metastasis1The Department of Magnetic Resonance Imaging, The First Affiliated Hospital of Zhengzhou University, Zhengzhou, China, 2Siemens Healthineers, Shanghai, China, 3Shanghai Key Laboratory of Magnetic Resonance, East China Normal University, Shanghai, China, 4The School of Software, Zhengzhou University, Zhengzhou, China
Synopsis
Differentiation of glioblastomas and solitary brain metastases is clinically crucial for the prescription of patient management and assessment of prognosis. However, indistinguishable signs between two tumors on routine MRI leads to a high misdiagnosis rate. Neurite orientation dispersion and density imaging (NODDI) can not only estimate the intricacy of neurites in vivo but also provide data to illuminate pathology. We developed a series of radiomics models of NODDI parameter maps, routine MRI, combined routine MRI, and combined NODDI parameter maps to compare their performance in the identification of two tumors. Finally, the combined NODDI radiomics model obtained the best performance.
Introduction
Glioblastomas (GBMs) and solitary brain metastases (SBMs) are among the most common malignant brain tumors. Their differentiation is an important clinical problem because the treatment strategy can substantially differ between the two [1]. Magnetic resonance imaging (MRI) is the main imaging method for the diagnosis of brain tumors. However, determining the differences between GBM and SBM is extremely challenging because they usually show similar signal characteristics and contrast enhancement patterns on routine MRI, leading to incorrect diagnosis in more than 40% of cases [2]. At present, three main tendencies occur in the identification of GBMs and SBMs. The first is the exploration of the differences among SBMs from different primary sites. The second is the improvement of the performance of imaging modalities, including routine and advanced MRI modes, such as neurite orientation dispersion and density imaging (NODDI). NODDI is a practical diffusion MRI technique that can be used to estimate the intricacy of neurites in vivo and performed on standard MRI scanners. The third tendency is the use of artificial intelligence technology to improve the capability of auxiliary diagnosis, such as radiomics. Radiomics has great potential in identifying GBMs and SBMs on routine MRI [3]. Nevertheless, whether the NODDI radiomics is superior to routine MRI radiomics in distinguishing GBMs and SBMs is still unknown. Therefore, we aimed to explore the performance of NODDI radiomics analysis in distinguishing GBMs and SBMs.Materials and Methods
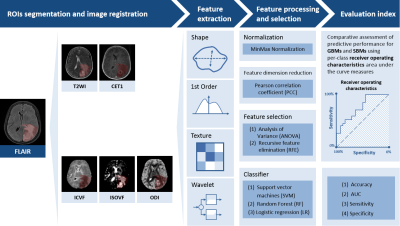
The institutional review board approved this prospective study, and informed consent was obtained from all patients. In November 2015 to April 2021, 101 patients were recruited for this study (50 GBMs and 51 SBMs). We randomly selected 81 cases as the training cohort (40/41 = positive/negative), whereas another 20 cases served as the independent testing cohort (10/10 = positive/negative). The inclusion criteria were as follows: 1) pathologically confirmed as GBMs; 2) pathologically confirmed or confirmed during follow-up as SBMs. The exclusion criteria were as follows: 1) lack of necessary MRI scans; 2) MRI scans with severe motion or susceptibility artifacts. All the patients underwent diffusion weighted imaging (DWI) and routine MRI examinations on a 3T MR scanner (MAGNETOM Prisma; Siemens Healthcare, Erlangen, Germany) with a 64-channel head-neck coil.The NODDI parameters, including intracellular volume fraction (ICVF), orientation dispersion index (ODI), and isotropic volume fraction (ISOVF), were calculated from DWI data using 5b values (500, 1000, 1500, 2000, and 2500 s/mm2) with 30 encoding directions for every b value by an in-house developed post-processing software called NeuDiLab (Based on DIPY, http://nipy.org/dipy). Figure 1 depicts the pipeline of data processing. Regions of interest were defined as the maximum abnormal signal area and outlined on the axial FLAIR image using ITK-SNAP (http://www.itksnap.org) software. Routine MRI maps and all NODDI parameter maps were spatially registered in accordance with FLAIR. FAE software was used for feature extraction, feature processing, feature selection, and model construction [4]. Each case extracted 851 radiomics features, including 18 first-order statistical features, 14 shape-based, 75 texture features of original and wavelet transformed images. Two feature selection methods and three classifiers were used to construct the radiomics prediction models on each of the routine MRI and NODDI parameter maps. In addition, a combined routine MRI model and a combined NODDI model were constructed. Fivefold cross-validation was used to prove the model performance, which was evaluated using receiver operating characteristic (ROC) curve, accuracy, the area under the ROC curve (AUC), sensitivity, and specificity on the testing cohort.Results
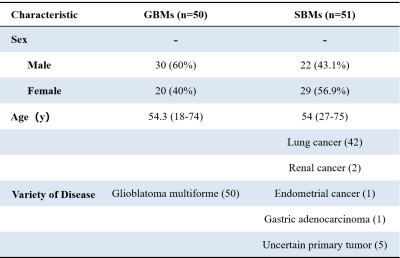
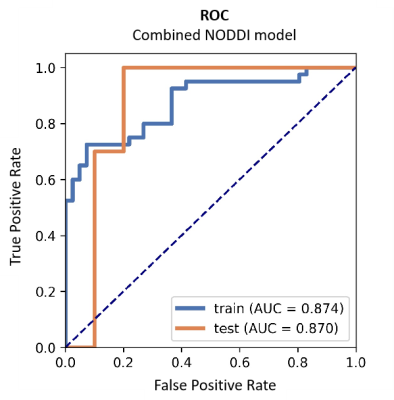
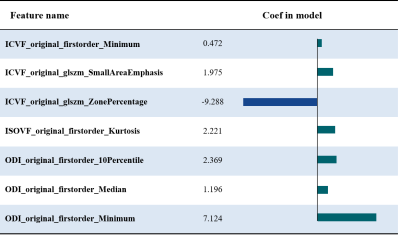
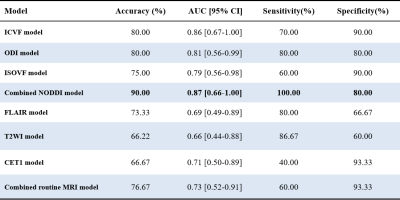
Table 1 summarizes the detailed clinical characteristics of the studied 101 patients. No significant difference was found between the two tumor groups for sex (P = 0.09) and age (P = 0.565).Figure 2 shows the best results obtained on the combined NODDI model. The model uses Recursive Feature Elimination (RFE) as feature selection and selects 7 key features to serve as the radiomics signature from the radiomics features (Table 2). The radiomics signature was used to build the predictive model by Logistic Regression (LR) classifier.Table 3 shows the performance of the two combined models, NODDI parametric map models, and routine MRI models. Our results indicated that the accuracy, AUC, sensitivity, and specificity for the combined NODDI model were 90.00%, 0.87, 100.00% and 80%, respectively. The values for the combined routine MRI models were 76.67%, 0.73, 60.00%, and 93.33%, respectively.Discussion
A recent study showed that ISOVF is the best NODDI parameter to distinguish between GBMs and SBMs, but it is worth noting that the study did not measure ODI [5]. Our results showed that ICVF and ODI are more useful for the differentiation of the two tumor types than ISOVF. The combined NODDI radiomics model of ISOVF, ICVF, and ISOVF can achieve better performance in identification, which is significantly better than that of the combined routine MRI radiomics models. NODDI simplifies the brain structure into three compartments per voxel: the intracellular space, extracellular space, and cerebrospinal fluid. NODDI can indicate more specific details on the microstructural changes in neurites than routine MRI, and these changes may be extracted and used as radiomics signature.Conclusion
Compared with routine MRI radiomics, NODDI radiomics analysis shows evident advantages in distinguishing GBMs and SBMs.Acknowledgements
This work was supported in part by the National Natural Science Foundation of China under Grant No. 81772009, and in part by Collaborative Innovation Major Project of Zhengzhou under Grant No. 20XTZX06013.References
[1] Kamimura K, Nakajo M, Yoneyama T, et al. Histogram analysis of amide proton transfer-weighted imaging: comparison of glioblastoma and solitary brain metastasis in enhancing tumors and peritumoral regions. Eur Radiol. 2019;29(8):4133-4140.
[2] Artzi M, Bressler I, Ben Bashat D. Differentiation between glioblastoma, brain metastasis and subtypes using radiomics analysis. J Magn Reson Imaging. 2019;50(2):519-528.
[3] Han Y, Zhang L, Niu S, et al. Differentiation Between Glioblastoma Multiforme and Metastasis From the Lungs and Other Sites Using Combined Clinical/Routine MRI Radiomics. Front Cell Dev Biol. 2021; 26(9):710461.
[4] Song Y, Zhang J, Zhang YD, et al. FeAture Explorer (FAE): A tool for developing and comparing radiomics models. PLoS One. 2020;15(8):e0237587.
[5] Fordham AJ, Hacherl CC, Patel N, et al. Differentiating Glioblastomas from Solitary Brain Metastases: An Update on the Current Literature of Advanced Imaging Modalities. Cancers (Basel). 2021;13(12):2960.
Figures