2977
Can MAP-MRI model be used for advance prediction of Grade II/III diffuse glioma genetics?
Peng Wang1, Yang Gao1, Qiong Wu1, Jinlong He1, Shenghui Xie1, Shaoyu Wang2, and Huapeng Zhang2
1Radiology, Affiliated Hospital of Inner Mongolia Medical University, Hohhot, China, 2Siemens Healthineers, Shanghai, China
1Radiology, Affiliated Hospital of Inner Mongolia Medical University, Hohhot, China, 2Siemens Healthineers, Shanghai, China
Synopsis
Although surgery is still the most effective treatment for diffuse glioma, patients with unresectable primary, recurrent, or multicentric illness have few options. In some cases, genotyping predictions may be useful for targeted therapy. We performed pretreatment MRI scans and genetic sequencing of tumor tissue, as well as correlation analysis, to see if essential molecular genetic events linked with diffuse glioma may be predicted in advance and noninvasively. The findings revealed that the MAP-MRI model can reliably predict tumor genotyping with high robustness and diagnostic validity, implying that it could be useful in the development of more standardized treatment.
Background and Purpose
Currently, there is no noninvasive method to effectively judge the genotype of diffuse gliomas.To explore whether mean apparent propagation diffusion magnetic resonance imaging (MAP-MRI) can predict the isocitrate dehydrogenase (IDH) 1/2 mutation and synchronous deletion of the short arm of chromosome 1 and long arm of chromosome 19 (1p/19q combined deletion) genotypes in grade II/III diffuse gliomas.Methods
In this study, 37 patients with pathologically confirmed diffuse glioma were included. According to the IDH and 1p/19q combined deletion status, the patients were divided into three groups [IDH wild type (IDHwt) and IDH mutant type (IDHmut), and IDHmut was divided into IDH mutant and 1p19q intact (IDHmut/1p19qint) and IDH mutant and 1p19q synchronous deletion (IDHmut/1p19qdel)] (Table 1). All the patients had undergone preoperative MRI on a 3T scanner (MAGNETOM Skyra; Siemens Healthcare, Erlangen, Germany) using a 32-channel head/neck coil, included conventional MRI and diffusion spectrum magnetic resonance imaging (DSI). The DSI sequence was obtained in the axial plane using a half q-space Cartesian grid sampling procedure under the following parameters: TR/TE = 7000/107 ms, FOV = 260 mm × 260 mm, GRAPPA = 2, layer thickness = 3.0 mm, voxel size = 2.2×2.2×3.0 mm3, 50 layers, maximum B value = 3000 s/mm2, and total scanning time = 15 min 40 s. The image was eddy and motion corrected in advance by DiffusionKit eddy tools. MAP-MRI parameters were calculated using NeuDiLab, a software developed in-house with Python based on the open-source tool DIPY (Diffusion Imaging in Python) [1]. The MAP-MRI parameters investigated included the mean square displacement (MSD), non-Gaussianity (NG), q-space inverse variance (QIV) and return to the origin probability (RTOP). The software program ITK-SNAP [2] was used to manually delineate the tumor parenchymal area. Ten regions of interest were selected to be placed on the solid portion of the tumor and ROIs were selected to avoid cystic degeneration, necrosis, hemorrhage, and calcification as much as possible, and the average of each diffusion parameter was used for analysis. Statistical software SPSS 24.0 (SPSS, Inc., Chicago, IL, USA) was used to analyze the data. Differences between demographic characteristics and tumor grading were analyzed by Chi-square test or one-way ANOVA. Correlations between quantitative parameters and genetic characteristics of diffuse gliomas were evaluated by independent samples t-test or Mann-Whitney U test. For parameters with P < 0.05, receiver operating characteristic curves (ROC) were constructed to assess diagnostic efficiency, and ROC stability was further assessed using k-fold cross-validation (K=5) by python.Results
The average MSD and QIV values of the IDHwt group were significantly lower than those of the IDHmut group (P<0.001), and the average NG and RTOP values were significantly higher than those in the IDHmut group (P<0.001). The average RTOP values of the IDHmut/1p19qdel group were lower than those of the IDHmut/1p19qint group (P<0.05) (Figure 1). The receiver operating characteristic curve was shown in Table 2, where QIV has the greatest diagnostic efficacy and remains high after cross-validation (Figure 2). The RTOP had low diagnostic efficacy in predicting 1p/19q combined deletion (AUC=0.73).Discussion and Conclusion
Our study found that MAP-MRI was effective in identifying IDH status in WHO grade II/III diffuse gliomas, thanks to MAP's ability to accurately estimate diffusion properties [3]. And by combining previous studies [4], MSD may be a novel diffusion indicator independent of tumor grade. MAP-MRI can effectively predict IDH 1/2 mutations and identify the 1p/19q combined deletion genotype of diffuse glioma, which can be a new imaging method to determine the genotype of diffuse glioma and help improve the clinical diagnosis and treatment process.Acknowledgements
No acknowledgement found.References
- Garyfallidis E, Brett M, Amirbekian B, et al. Dipy, a library for the analysis of diffusion MRI data. Front Neuroinform 2014;8:8.2.
- Yushkevich PA, Piven J, Hazlett HC, et al. User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage 2006;31:1116-1128.3.
- Ning L, Laun F, Gur Y, et al. Sparse Reconstruction Challenge for diffusion MRI: Validation on a physical phantom to determine which acquisition scheme and analysis method to use?. Med Image Anal. 2015;26(1):316-331. doi:10.1016/j.media.2015.10.0124.
- Wang P, Weng L, Xie S, et al. Primary application of mean apparent propagator-MRI diffusion model in the grading of diffuse glioma. Eur J Radiol 2021;138:109622.
Figures
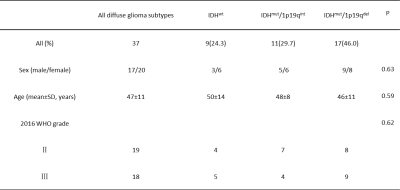
Table 1. Patient demographics. IDH : isocitrate dehydrogenase, 1p/19q : synchronous deletion of the short arm of chromosome 1 and long arm of chromosome 19, IDHwt : IDH wild-type, IDHmut/1p19qint : IDH mutant and 1p19q intact, IDHmut/1p19qdel : IDH mutant and 1p19q synchronous deletion, WHO : World Health Organization.
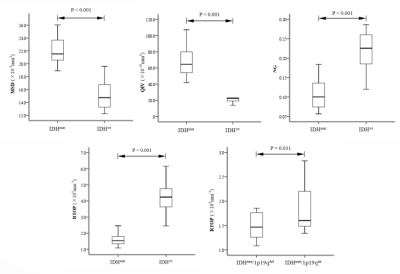
Figure 1. Boxplot shows differences in the MAP-MRI parameters between World Health Organization grade II/III glioma molecular subtypes in the study sample (9 wild-type isocitrate dehydrogenase [IDH; IDHwt]; 28 IDH mutation [IDHmut; 11 IDHmut/1p19qint, and 17 IDHmut/1p19qdel). MSD : mean square displacement, QIV : q-space inverse variance, NG : non-Gaussianity, RTOP : return to the origin probability.
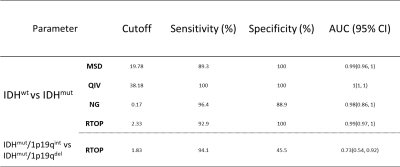
Table 2. ROC analysis of MAP-MRI parameters for predicting diffuse glioma genotypes. IDH : isocitrate dehydrogenase, 1p/19q : synchronous deletion of the short arm of chromosome 1 and long arm of chromosome 19, IDHwt : IDH wild-type, IDHmut/1p19qint : IDH mutant and 1p19q intact, IDHmut/1p19qdel : IDH mutant and 1p19q synchronous deletion, WHO : World Health Organization. MSD : mean square displacement, QIV : q-space inverse variance, NG : non-Gaussianity, RTOP : return to the origin probability.
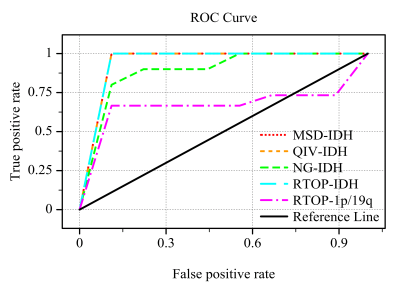
Figure 2. Diagnostic efficacy of MAP-MRI parameters after k-fold cross-validation (K=5). MSD : mean square displacement, QIV : q-space inverse variance, NG : non-Gaussianity, RTOP : return to the origin probability; -IDH : identify IDH status, -1p/19q : identify 1p/19q combined deletion status.
DOI: https://doi.org/10.58530/2022/2977