2951
Stratified Parkinsonism classification based on multi-modality MRI1Academy for Engineering & Technology, Fudan University, Shanghai, China, 2Department of Radiology, Huashan Hospital, Shanghai, China, 3Institute of Functional and Molecular Medical Imaging, Fudan University, Shanghai, China, 4Department of Neurology, Huashan Hospital, Shanghai, China, 5GE Healthcare, Beijing, China
Synopsis
For differential diagnosis between PD and atypical parkinsonisms, we designed a stratified automated method based on SVM and multi-modality MRI, including T1WI, QSM and DTI. 163 patients (96 PD, 27 MSA, 40 PSP) and 65 healthy controls were recruited. Features including volume, cortical thickness, magnetic susceptibility, FA and MD of 124 ROIs were calculated for SVM classification. The result showed that SVM classification based on susceptibility enabled accurate differentiation of patients with parkinsonian disorders and controls, and classification of PD, MSA and PSP was allowed by using T1 and DTI.
Introduction
The diagnosis of Parkinson's disease (PD) is still difficult especially in early PD patients[1]. In order to improve the accuracy of clinical PD diagnosis, it is necessary to differentiate PD and atypical parkinsonism, mainly including progressive supranuclear palsy (PSP) and multiple system atrophy (MSA). Diagnosis of PD and atypical parkinsonism mainly relies on clinical criteria[2-5], but clinical standards are generally difficult to make a correct diagnosis. A clinical research reported an error rate of 10%-30% in the diagnosis of early PD[6], indicating that clinical parkinsonism classification remains a diagnostic challenge.Magnetic resonance imaging (MRI) is an important method for studying brain structure, and has been widely used in the differential diagnosis of PD, MSA and PSP. However, most of recent studies used only susceptibility and DTI parameters[7-9], or only volumetry data[10-12]. Most of them used a single modality, and a limited number of ROIs. In addition, the ROIs were often drawn manually, which was less efficient. Since previous studies have also induced machine learning methods in parkinsonism differentiation[7, 13, 14], in order to discover more regions related to symptoms, and to make full use of the advantage of machine learning of dealing with a large number of features, obtaining and using more ROIs by an automatic method is necessary.
In this study, we aim to develop an automated stratification method based on support vector machine (SVM) and multi-modality MRI, including T1-weighted imaging (T1WI), quantitative susceptibility mapping (QSM) and diffusion tensor imaging (DTI), and evaluate its performance in differential diagnosis of PD, MSA and PSP.
Methods
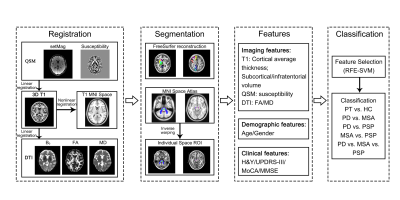
The overall procedure of this study is shown in Fig. 1. 163 patients (96 PD, 27 MSA, 40 PSP) and 65 healthy controls (HCs) were recruited in this study. All MRI examinations were performed on a 3T MRI scanner (GE Healthcare, Milwaukee, WI). 3D T1WI, QSM and DTI images were acquired from all participants. Data from all participants were divided into a training cohort and a test cohort in a ratio of 4:1. 68 cortical and 56 subcortical regions of interest (ROI) were obtained by automatic segmentation using FreeSurfer reconstruction and atlas registration. Average thickness of cortical ROI, volume of subcortical ROI, average susceptibility, FA, MD of each ROI were calculated for each subject as features for classification.The classification used SVM with a radial basis function (RBF) kernel. The input matrices were as follows: T1, QSM, FA, MD, DTI (FA+MD), T1+QSM, T1+DTI, DTI+QSM, T1+DTI+QSM, and clinical features (age, gender, UPDRS-III, H-Y, MoCA and MMSE). Recursive feature elimination SVM (RFE-SVM) with correlation bias reduction was used for feature selection. The input was first normalized, and the SVM hyperparameters were optimized by a grid search with 5-fold cross-validation in the training cohort. Area under the curve (AUC), accuracy, sensitivity, and specificity were calculated to evaluate the performance of SVM algorithm.
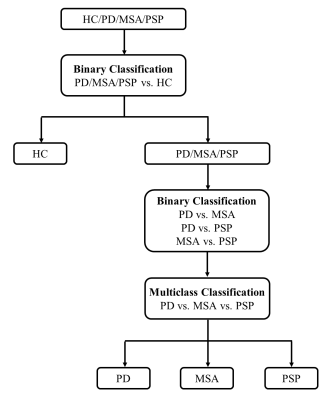
The classification used a stratified strategy which is shown in Fig. 2. First, patients and HCs were classified using a binary classification. Then the three groups of patients, PD, MSA and PSP, were classified in a “one-versus-one” way by using three binary classifications, PD vs MSA, PD vs PSP, and MSA vs PSP with the best performance. Accuracy, specificity and sensitivity was then calculated to evaluate the performance of multiclass classification.
Results
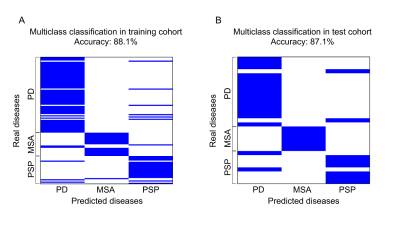
In the test cohort, susceptibility performed best in the classification of patients–HC (accuracy: 0.775, specificity: 0.833, sensitivity: 0.750). The most relevant feature between patients and HCs occurred in susceptibility of substantia nigra pars reticulate (SNr). The combination of T1 and DTI features showed the best performance in the classification of PD–MSA (accuracy: 0.958, specificity: 0.941, sensitivity: 1.000) and MSA–PSP (accuracy: 1.000, specificity: 1.000, sensitivity: 1.000). T1 features performed best in the classification of PD–PSP (accuracy: 0.929, specificity: 0.950, sensitivity: 0.875). PD, MSA and PSP differed mainly in T1, FA and MD values in areas of pons, substantia nigra pars compacta (SNc) and pontine crossing tract (PCT). Results of multiclass SVM multiclass classification of PD, MSA and PSP are shown in Fig. 3. Accuracy was 88.1% in the training cohort and 87.1% in the test cohort. Specificity and sensitivity were calculated for each group by regarding one group as positive and the other two as negative, which were 0.881 and 0.881 for PD, 0.988 and 0.586 for MSA, 0.916 and 0.769 for PSP in the training cohort, and 0.857 and 0.882 for PD, 1.000 and 0.600 for MSA, 0.913 and 0.750 for PSP in the test cohort.Discussion and Conclusion
Differential diagnosis of PD and atypical parkinsonism is still difficult, and most of recent related researches were limited in modality and methods. In our study, a stratified automated method based on SVM classification and multi-modality MRI was designed and evaluated, and the results proved that SVM classification based on susceptibility enabled accurate differentiation between patients with parkinsonian disorders and controls, and T1 and DTI parameters could be valuable imaging biomarkers for the classification of PD, MSA and PSP.Acknowledgements
This work was supported by National Natural Science Foundation of China [82102132] and Science and Technology Commission of Shanghai Municipality [20S31904300].References
1. Rizzo G, Copetti M, Arcuti S, et al. Accuracy of clinical diagnosis of Parkinson disease: A systematic review and meta-analysis. Neurology 2016, 86(6): 566-576.
2. Gilman S, Wenning GK, Low PA, et al. Second consensus statement on the diagnosis of multiple system atrophy. Neurology 2008, 71(9): 670-676.
3. Höglinger GU, Respondek G, Stamelou M, et al. Clinical diagnosis of progressive supranuclear palsy: The movement disorder society criteria. Movement Disorders 2017, 32(6): 853-864.
4. Wenning GK, Colosimo C, Geser F, et al. Multiple system atrophy. The Lancet Neurology 2004, 3(2): 93-103.
5. Williams DR, Lees AJ. Progressive supranuclear palsy: clinicopathological concepts and diagnostic challenges. The Lancet Neurology 2009, 8(3): 270-279.
6. Jankovic J, Rajput AH, McDermott MP, et al. The evolution of diagnosis in early Parkinson disease. Parkinson Study Group. Arch Neurol 2000, 57(3): 369-372.
7. Cherubini A, Morelli M, Nisticó R, et al. Magnetic resonance support vector machine discriminates between Parkinson disease and progressive supranuclear palsy. Movement Disorders 2014, 29(2): 266-269.
8. Sjöström H, Granberg T, Westman E, et al. Quantitative susceptibility mapping differentiates between parkinsonian disorders. Parkinsonism & Related Disorders 2017, 44: 51-57.
9. Prodoehl J, Li H, Planetta PJ, et al. Diffusion tensor imaging of Parkinson's disease, atypical parkinsonism, and essential tremor. Movement Disorders 2013, 28(13): 1816-1822.
10. Paviour DC, Price SL, Jahanshahi M, et al. Regional brain volumes distinguish PSP, MSA-P, and PD: MRI-based clinico-radiological correlations. Movement Disorders 2006, 21(7): 989-996.
11. Sjostrom H, Granberg T, Hashim F, et al. Automated brainstem volumetry can aid in the diagnostics of parkinsonian disorders. Parkinsonism Relat Disord 2020, 79: 18-25.
12. Scherfler C, Göbel G, Müller C, et al. Diagnostic potential of automated subcortical volume segmentation in atypical parkinsonism. Neurology 2016, 86(13): 1242-1249.
13. Haller S, Badoud S, Nguyen D, et al. Differentiation between Parkinson disease and other forms of Parkinsonism using support vector machine analysis of susceptibility-weighted imaging (SWI): initial results. European Radiology 2013, 23(1): 12-19.
14. Chougar L, Faouzi J, Pyatigorskaya N, et al. Automated Categorization of Parkinsonian Syndromes Using Magnetic Resonance Imaging in a Clinical Setting. Movement Disorders 2021, 36(2): 460-470.
Figures