2905
Improved MP-PCA denoising through tensor generalization1Center of Functionally Integrative Neuroscience (CFIN) and MINDLab, Aarhus University, Aarhus, Denmark, 2Department of Physics and Astronomy, Aarhus University, Aarhus, Denmark, 3Champalimaud Research, Champalimaud Centre for the Unknown, Lisbon, Portugal
Synopsis
A popular SNR-boosting method in MRI is denoising based on principal component analysis with automated rank estimation by exploiting the Marchenko-Pastur distribution of noise singular values (MP-PCA). MP-PCA operates by reshaping data-patches into matrices to discriminate signal from noise using random matrix theory. Here, we generalize MP-PCA to exploit tensor-structured data arising in, e.g., multi-contrast or multicoil acquisitions, without introducing new assumptions. As proof of concept, we demonstrate a substantial increase in denoising performance in a multi-TE DKI dataset, in particular for small sliding windows. This is beneficial especially in cases of rapidly varying contrast or spatially varying noise.
Introduction
MP-PCA1,2 denoising is based on principal component analysis (PCA) and exploits the Marchenko-Pastur (MP) distribution of noise singular values for rank estimation. MP-PCA has become very popular for SNR-boosting with negligible signal loss or spatial smoothing3-6 because of its effectiveness and objective rank estimation.Information redundancy in MRI acquisitions (e.g. in directions in DTI or TE in T2 mapping) make typical MRI data well described by a low-rank approximation suitable for MP-PCA denoising. However, in many cases, multiple dimensions with redundancy exist, which opens opportunities for improved denoising. Here, we generalize matrix MP-PCA to tensor-structured data (tensor MP-PCA) and exploit the redundancy in multiple dimensions by applying MP rank-reduction to the higher-order singular value decomposition (HOSVD) recursively, without introducing new assumptions. Tensor MP-PCA performs as well or better than matrix MP-PCA in general, and we demonstrate substantial improvements in tensor-structure scenarios. Through objective rank estimation, recursivity, and shrinkage4,8, tensor MP-PCA also outperforms classical HOSVD approaches7.
Theory
Matrix MP-PCAMP-PCA1,2 denoising applies a “sliding window” over voxels and denoises the resulting patches independently. Each patch forms a matrix $$$X$$$ with voxels as rows and one or more modalities (i.e. diffusion, relaxation, fMRI) as columns. SVD yields $$X=USV^\dagger$$ then noise components are removed based on the MP distribution of noise singular values (assuming iid noise with zero mean). Thereby estimating $$$\sigma^2$$$ and the signal subspace onto which $$$X$$$ is projected to yield a denoised $$$\tilde{X}$$$ $$\tilde{X}=\tilde{U}\tilde{U}^{\dagger}X=\tilde{U}\tilde{S}\tilde{V}^\dagger$$ where tilde signifies that noise components have been removed. Some improvement is attained by applying optimal shrinkage4,8.
Tensor MP-PCA
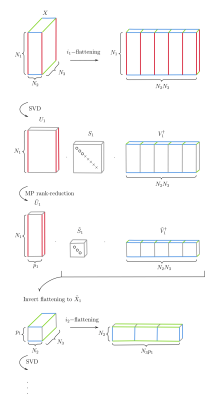
Given multiple dimensions, each patch can form a tensor. Assume for concreteness but without loss of generality that each patch is a three-index tensor $$$X_{i_1i_2i_3}$$$ (see Fig. 1) – for instance in dMRI with $$$i_1$$$ for voxels, $$$i_2$$$ for directions, and $$$i_3$$$ for b-values. Take $$$X$$$’s $$$i_1$$$-flattening $$$X_{i_1(i_2i_3)}$$$ (the parenthesis signifies index concatenation) and apply SVD and MP rank-reduction yielding $$$\tilde{U}\tilde{S}\tilde{V}^\dagger$$$. Define tensor $$$\tilde{X}_1\equiv\tilde{U}_1^{\dagger}X=[\tilde{S}_1\tilde{V}_1^\dagger]$$$, where it is implicit that $$$\tilde{U}^\dagger$$$ acts on index $$$i_1$$$ and $$$[\cdots]$$$ signifies inverting the previous flattening. $$$\tilde{X}_1$$$ retains $$$X$$$'s noise properties, since $$$\tilde{U}_1$$$ is unitary. Applying SVD and rank-reduction to the $$$i_2$$$-flattening of $$$\tilde{X}_1$$$ yields $$$\tilde{U}_2$$$ and $$$\tilde{X}_2\equiv\tilde{U}_2^{\dagger}\tilde{X}_1=[\tilde{S}_2\tilde{V}_2^\dagger]$$$ and so on. The recursive procedure stops at the last index after which $$\tilde{X}=\tilde{U}_1\tilde{U}_2\tilde{U}_3[\tilde{S}_3\tilde{V}_3^\dagger]$$ This reduces to matrix MP-PCA for matrices resulting in equal or better performance. It outperforms classical HOSVD approaches7, for which the rank has been determined a priori, through objective rank estimation and the recursive approach, which removes noise components at step $$$n$$$ before proceeding to $$$n+1$$$ - improving the $$$U_{n+1}$$$, $$$X_{n+1}$$$ estimates - rather than estimating all $$$U$$$ before rank-reduction. Furthermore, shrinkage is applicable to the last $$$\tilde{S}_n$$$.
Methods
All experiments were preapproved by the competent institutional and national authorities and were carried out in accordance with European Directive 2010/63.For proof of concept, we employ a mouse brain dMRI dataset recorded at 16.4T using a microimaging probe with a multi-spin-echo sequence (FOV 10x8mm2, matrix 100x80, 27 slices, in-plane resolution 0.1x0.1mm2, slice-thickness 0.5mm, pulse width 3.5ms, pulse separation 5.6ms, 6 b-shells from 0.5 to 3ms/μm2, 20 directions, and repeated for 10 TE from 10.7 to 35ms). Matrix and tensor MP-PCA are compared using the data and a numerical phantom created by generating a DKI signal (parameters from a DKI fit to actual data) for each TE and adding Gaussian noise with an SNR ($$$S_0/\sigma$$$) of twenty, ten b-shells, thirty-two directions, and one/ten TEs. Performance is quantified using the root-mean-squared-error (RMSE) between the denoised and ground truth images
Results and discussion
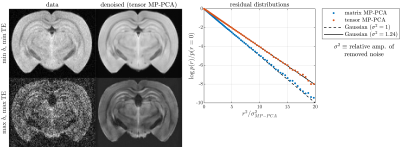
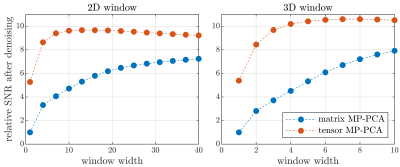
Fig. 1 shows data before and after applying tensor MP-PCA for the highest and lowest SNR scenarios. Residual distributions for tensor and matrix techniques are shown to be approximately Gaussian indicating that negligible signal variance is removed. For this dataset, tensor MP-PCA removes 24% more noise than matrix MP-PCA. Performance depends on several properties such as SNR, with better relative performance in low-SNR scenarios as suggested by the numerical phantom (Fig. 2).Fig. 2 compares performance on multi-TE phantom data for various sliding window sizes. Tensor MP-PCA outperforms matrix MP-PCA for any window size, consistent with containing matrix PCA as a special case. Tensor MP-PCA reaches optimal performance at smaller patches, making it better suited for spatially varying noise or rapidly varying contrast. Additionally, the smaller patches reduces computation time.
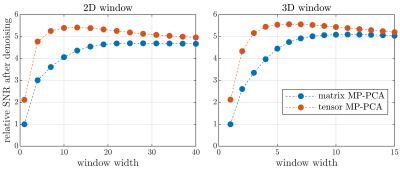
Fig. 3 compares performance on single-TE phantom data. Comparing Fig. 2 and 3 it is apparent that the tensor-structure from multiple TEs benefits tensor MP-PCA substantially. This opens several perspectives on particularly favorable applications such as multicoil3 or multimodality acquisitions9-12. We also note that MRI data is inherently tensor-structured in terms of voxels with x, y, z indices. For the data considered here, however, we found that combining voxels in one index performs near optimal. Nevertheless, it is plausible that retaining the voxel tensor-structure could increase performance in other scenarios7.
Conclusion
We generalized MP-PCA to multidimensional data achieving improved performance in such scenarios. Tensor MP-PCA works well for smaller patches increasing speed and suitability to rapidly varying contrast and spatially varying noise. Since matrix MP-PCA is a special case of tensor MP-PCA and no new assumptions are introduced, tensor MP-PCA can be directly adopted wherever MP-PCA is applied.Acknowledgements
The authors thank Martin Mikkelsen for support with the design and creation of illustrations.
SJ and JO are supported by the Danish National Research Foundation (CFIN), and the Danish Ministry of Science, Innovation, and Education (MINDLab). Additionally, JO are supported by the VELUX Foundation (ARCADIA, grant no. 00015963). NS was supported in part by the European Research Council (ERC) (agreement No. 679058). AI is supported by ”la Caixa” Foundation (ID 100010434) and European Union’s Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement No 847648, fellowship code CF/BQ/PI20/11760029. The authors acknowledge the vivarium of the Champalimaud Centre for the Unknown, a facility of CONGENTO which is a research infrastructure co-financed by Lisboa Regional Operational Programme (Lisboa 2020), under the PORTUGAL 2020 Partnership Agreement through the European Regional Development Fund (ERDF) and Fundação para a Ciência e Tecnologia (Portugal), project LISBOA-01-0145-FEDER-022170.
References
1. Veraart et al. Diffusion MRI noise mapping using random matrix theory. Magn Reson Med. 2016
2. Veraart et al. Denoising of diffusion MRI using random matrix theory. NeuroImage. 2016
3. Lemberskiy et al. Achieving sub-mm clinical diffusion MRI resolution by removing noise during reconstruction using random matrix theory. ISMRM proceedings. 2019
4. Cordero-Grande et al. Complex diffusion-weighted image estimation via matrix recovery under general noise models. NeuroImage. 2019
5. Does et al. Evaluation of principal component analysis image denoising on multi-exponential MRI relaxometry. Magn Reson Med. 2019
6. Zhao et al. Detail-Preserving Image Denoising via Adaptive Clustering and Progressive PCA Thresholding. IEEE Acess. 2018
7. Brender et al. Dynamic Imaging of Glucose and Lactate Metabolism by 13 C-MRS without Hyperpolarization. Scientific Reports. 2019
8. Gavish et al. Optimal Shrinkage of Singular Values. IEEE Trans Inf Theory. 2017
9. Veraart et al. TE dependent Diffusion Imaging (TEdDI) distinguishes between compartmental T2 relaxation times. NeuroImage. 2018
10. Lampinen et al. Towards unconstrained compartment modeling in white matter using diffusion-relaxation MRI with tensor-valued diffusion encoding. Magn Reson Med. 2020
11. Kleban et al. Strong diffusion gradients allow the separation of intra- and extra-axonal gradient-echo signals in the human brain. NeuroImage. 2020
12. Shemesh and Jespersen et al. Conventions and nomenclature for double diffusion encoding NMR and MRI. Magn Reson Med. 2015
Figures