2846
NODDI-FAST derived parameters are echo-time independent1Magnetic Resonance Physics of Aging and Dementia Unit, National Institute on Aging, NIH, Baltimore, MD, United States
Synopsis
In a previous work, we introduced the NODDI-FAST approach to address issues regarding overestimated CSF and neurite density (NDI) fractions in white matter seen with the original NODDI approach. However, in both NODDI-FAST and NODDI signal models, the compartment-specific T2 relaxations are not considered. Therefore, derived parameter estimates, especially NDI, could be dependent on echo time (TE). Here, we show that, as expected, ODI derived values using either NODDI or NODDI-FAST are TE-independent. We also confirm that NDI derived values using NODDI are TE-dependent. More importantly, we show that NDI derived values using NODDI-FAST are TE-independent.
Introduction
Neurite orientation dispersion and density imaging (NODDI) provides measures of the orientation dispersion index (ODI) and the neurite density index (NDI) (1). NODDI is based on a tricomponent diffusion model corresponding to intracellular, extracellular and CSF water compartments. However, this signal model intrinsically assumes that all compartments exhibit similar T2 values; this assumption leads to overestimated CSF and NDI in white matter (WM) (1-3). Bouyagoub and colleagues have suggested rescaling CSF fraction using predetermined T2 values of the CSF and intra/extracellular water compartments (3), while Gong and colleagues proposed a multi-echo time (mTE)-NODDI approach incorporating several NODDI scans performed at different TEs (2). These advanced approaches have led to lower CSF and plausible NDI values in WM. Nevertheless, they drastically extend the total acquisition time, making them impractical in clinical settings. Recently, we have proposed a new variant of NODDI, named, NODDI-FAST (4). In NODDI-FAST, the CSF fraction is derived using the FAST algorithm (5, 6) (from a structural image acquired within a few seconds), and provided as an input (i.e., known) value in each voxel instead of being estimated along with NDI and ODI. This fitting of a bicomponent model, incorporating only the intracellular and extracellular water compartments, replaces the original tricomponent model used in NODDI, leading to NDI values within a physiological plausible range (3, 4). However, as in NODDI, the signal model of NODDI-FAST does not account for compartment-specific T2 relaxations. However, it is known that the T2 of the intracellular and extracellular compartments are very similar (7-11). Therefore, it is expected that, unlike NODDI, derived parameters from NODDI-FAST are TE-independent.Methods
Data Acquisition Diffusion-weighted (DW) images were acquired from the brain of a participant at four different TEs of 78, 90, 100 or 120 ms using a single-shot EPI sequence with repetition time (TR) of 10 s, three b-values of 0, 700 and 2000 s/mm2, with the latter two encoded in 32 diffusion-weighting gradient directions, field-of-view (FoV) of 240 mm × 208 mm × 150 mm, acquisition matrix of 120 × 102 × 75, acquisition voxel size of 2 mm × 2 mm × 2 mm, SENSE factor of 2, and acquisition time of ~16 min. Finally, a 3D spoiled gradient recalled echo image was acquired with flip angle of 10°, TR of 5 ms, TE of 1.46 ms, FoV of 240 mm × 208 mm × 150 mm, matrix size of 120 × 102 × 75, acquisition voxel size of 2 mm × 2 mm × 2 mm, SENSE factor of 2, and acquisition time of ~30 s. This T1-weigthed image was used to calculate the CSF fraction, using the FAST algorithm (5, 6), and was provided as an input in NODDI-FAST for the determination of NDI and ODI (4).NDI and ODI mapping and analysis
For each NODDI dataset acquired at a given TE, the corresponding DW images were registered to the b0 image and corrected for motion and eddy current distortion artefacts using the Artefact Correction in Diffusion MRI toolbox (12). Then, the NDI and ODI maps were generated using NODDI or NODDI-FAST (1, 4). Finally, for sake of illustration, ODI and NDI values were calculated in a region-of-interest covering a large WM region.
Results and Discussion
Figure 1 shows representative NDI maps derived using NODDI or NODDI-FAST, at each of the four TEs studied, for a representative axial slice. Visual inspection indicates that derived NDI maps using NODDI exhibit unrealistic values exceeding 70% in several brain structures, while NDI values derived from NODDI-FAST are much lower and within the expected physiological range. These results agree with Bouyagoub et al. observations (3). Importantly, these results indicate that the NDI values derived using NODDI increase with increasing TE, while the NDI values derived using NODDI-FAST are similar across TEs. Our quantitative analysis shown in Figure 2 confirms these observations. Aside from providing a clinically practical method to map NDI that is indepentent of the choice of TE, our analysis also highlights the fact that the NDI independence of TE in NODDI-FAST is an indicator that the T2s of the intracellular and extrcellular water compartements are very similar. While this agrees with the extensive relaxometry-based observations (7-11), our results contradict Gong and colleagues’ assumption of different T2s between the intracellular and extracellular water compartements (2). The mTE-NODDI, which involves acquiring several NODDI datasets at different TEs, as proposed by Gong and colleagues, incoporates several pre- and post-processing steps and strong prior limits imposed on the T2 values of each water compartement. This could lead to an artifical determination of these T2 values; this is commonly seen in multicompnent analyses (13, 14).Finally the ODI values derived using NODDI or NODDI-FAST were virtually similar and indepentent of TE (Figs. 3-4), as expected. Indeed, the calculation of ODI does not incorporate signal fractions and thus remains insensitive to differences in the CSF fraction.
Conclusions
Unlike NODDI, NODDI-FAST derived parameters exhibit no dependence on TE. NODDI-FAST allows accurate determination of the neurite density and dispersion in a clinically feasible acquisition time, and will potentially permit comparison of results from multi-shell diffusion datasets acquired at different TEs.Acknowledgements
This work was supported by the Intramural Research Program of the National Institute on Aging of the National Institutes of Health.
References
1. Zhang H, Schneider T, Wheeler-Kingshott CA, Alexander DC. NODDI: practical in vivo neurite orientation dispersion and density imaging of the human brain. NeuroImage. 2012;61(4):1000-16.
2. Gong T, Tong Q, He H, Sun Y, Zhong J, Zhang H. MTE-NODDI: Multi-TE NODDI for disentangling non-T2-weighted signal fractions from compartment-specific T2 relaxation times. NeuroImage. 2020;217:116906.
3. Bouyagoub S, Dowell N, Hurley S, TC. W, M. C. Overestimation of CSF fraction in NODDI: possible correction techniques and the effect on neurite density and orientation dispersion measures. ISMRM; Singapore2016.
4. Maryam A, Wenshu Q, Matthew K, Curtis T, Zhaoyuan G, Mustapha B. Neurite dispersion and density across the adult lifespan investigated using a modified NODDI approach. ISMRM2021.
5. Zhang Y, Brady M, Smith S. Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE transactions on medical imaging. 2001;20(1):45-57.
6. Jenkinson M, Beckmann CF, Behrens TE, Woolrich MW, Smith SM. FSL. NeuroImage. 2012;62(2):782-90.
7. Kalantari S, Laule C, Bjarnason TA, Vavasour IM, MacKay AL. Insight into in vivo magnetization exchange in human white matter regions. Magnetic resonance in medicine. 2011;66(4):1142-51.
8. Whittall KP, MacKay AL, Graeb DA, Nugent RA, Li DK, Paty DW. In vivo measurement of T2 distributions and water contents in normal human brain. Magnetic resonance in medicine. 1997;37(1):34-43.
9. Does MD. Inferring brain tissue composition and microstructure via MR relaxometry. NeuroImage. 2018;182:136-48.
10. Bouhrara M, Spencer RG. Improved determination of the myelin water fraction in human brain using magnetic resonance imaging through Bayesian analysis of mcDESPOT. NeuroImage. 2016;127:456-71.
11. Alonso-Ortiz E, Levesque IR, Pike GB. MRI-based myelin water imaging: A technical review. Magnetic resonance in medicine. 2015;73(1):70-81.
12. Mohammadi S, Möller HE, Kugel H, Müller DK, Deppe M. Correcting eddy current and motion effects by affine whole‐brain registrations: Evaluation of three‐dimensional distortions and comparison with slicewise correction. Magnetic Resonance in Medicine. 2010;64(4):1047-56.
13. West DJ, Teixeira RPAG, Wood TC, Hajnal JV, Tournier J-D, Malik SJ. Inherent and unpredictable bias in multi-component DESPOT myelin water fraction estimation. NeuroImage. 2019;195:78-88. 14. Bouhrara M, Reiter DA, Celik H, Fishbein KW, Kijowski R, Spencer RG. Analysis of mcDESPOT- and CPMG-derived parameter estimates for two-component nonexchanging systems. Magnetic resonance in medicine. 2016;75(6):2406-20.
Figures
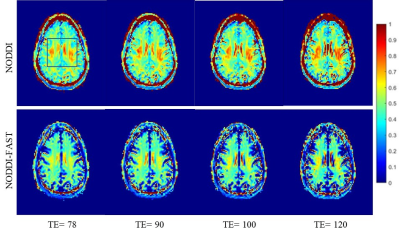
Figure 1. NDI maps derived using NODDI or NODDI-FAST from DW data acquired at four different SNRs. Results are shown for a representative slice. Visual inspection indicates that NDI values derived using NODDI increase with TE, while remaining constant using NODDI-FAST.
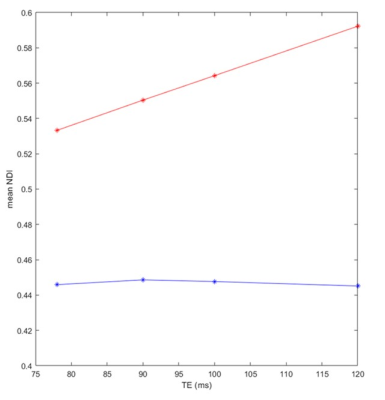
Figure 2. Mean NDI values, calculated from a large WM region, derived using NODDI (red) or NODDI-FAST (blue) as a function of TE. It is readily seen from this quantitative analysis that the NDI values derived using NODDI increase with TE, while remain constant using NODDI-FAST; this agrees with Fig. 1.
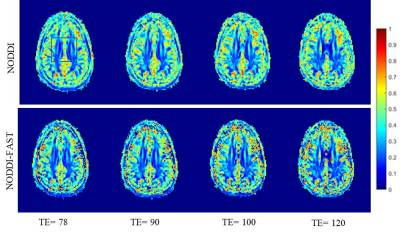
Figure 3. ODI maps derived using NODDI or NODDI-FAST from DW data acquired at four different SNRs. Results are shown for a representative slice. Visual inspection indicates that, for both NODDI and NODDI-FAST approaches, ODI values are similar for all TEs.
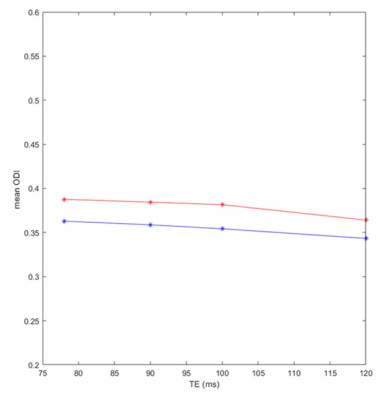
Figure 4. Mean ODI values, calculated from a large WM region, derived using NODDI (red) or NODDI-FAST (blue) as a function of TE. It is readily seen from this quantitative analysis that the ODI values derived using either NODDI or NODDI-FAST are relatively constant as a function of TE; this agrees with Fig. 3.