2836
A Follow-up Study On Prospective Para-Clinical Use by Residents of a Re-calibrating Automated Deep Learning System for Prostate Cancer Detection1Department of Radiology, German Cancer Research Center, Heidelberg, Germany, 2Department of Urology, University Hospital Heidelberg, Heidelberg, Germany, 3Institute of Pathology, University of Heidelberg Medical Center, Heidelberg, Germany
Synopsis
Previously validated fully-automatic detection of prostate cancer by CNNs requires further prospective validation. Para-clinical case-by-case prospective prostate MRI assessment by residents was performed both before and after review of CNN probability maps superimposed on T2w images. A previously and retrospectively validated self-parametrizing nnUNet-architecture CNN trained on more than 1000 voxel-wise annotated prostate MRIs achieved ROC AUC of 0.89. Residents did not substantially change their assessment both at PI-RADS>=3 and >=4 decisions, however achieved excellent working points, indicating success of high reading capability conveyed at an expert center.
INTRODUCTION
We have recently validated a deep learning (DL) convolutional neural network (CNN)-based fully-automated system for detection and segmentation of suspicious lesions on prostate MRI utilizing the nnUNet architecture (1). The now self-calibrating nnUNet approach demonstrated similar performance compared to an earlier hand-adjusted U-Net model (PROUNET) (2) which had been trained with 316 exams. Increasing the training set size to more than 1000 exams provided improvement to earlier models and similar performance to clinical radiologist assessment. The retrospectively well-validated performance requires further prospective validation. A pilot study on para-clinical case-by-case application of PROUNET has demonstrated susceptibility to calibration shifts over time (data not shown), suggesting that continuous sliding-window threshold calibration as previously proposed should be applied (3). The aim of this study was to evaluate prospective para-clinical use of an updated CNN DL system, utilizing the nnUnet architecture and continuous threshold re-calibration, by radiology residents and compare performances to clinical radiologist assessment.METHODS
140 mpMRI examinations of patients receiving subsequent extended systematic and targeted transperineal MR/TRUS fusion biopsy between October 2020 and June 2021 were included. Clinically significant prostate cancer (sPC) was defined as ISUP Grade Group >=2. 51 patients harbored clinically significant PCa (sPC). On a case-by-case basis, bi-parametric MRI data consisting of T2-weighted, DWI b=1500 s/mm2 and corresponding ADC maps were exported and automatically analyzed by a processing pipeline performing DWI/ADC to T2W image co-registration and image segmentation by a CNN trained with more than 1000 exams with full voxel-wise annotation of radiologist-identified lesions performed between 2015 and before October 2020. The CNN utilized a sliding-windows threshold re-calibration to maintain performance as proposed before (3). CNN-derived probability maps were presented as colormaps superimposed on anatomical T2-weigted scans. 16 radiology residents read between 1 and 19 examinations in a para-clinical setup, in which research re-reads of MR exams were performed without interaction with the clinical team and blinded to clinical information. Readers recorded a PI-RADS score for each detected lesion and performed two reads, one before and one after review of the CNN results. Clinical MR reads were later collected retrospectively and compared with resident assessments and CNN performance using the McNemar test.RESULTS
CNN achieved ROC AUC of 0.89. Clinical radiologist assessment had sensitivity/specificity of 1.00/0.12 at PI-RADS>=3 and 0.96/0.60 at PI-RADS >=4, respectively. Residents had 0.96/0.31 before and 0.96/0.31 after CNN review for PI-RADS>=3 and 0.86/0.64 before and 0.88/0.63 after CNN review for PI-RADS>=4. CNN had sensitivity/specificity of 0.96/0.24 at PI-RADS>=3 and 0.94/0.47 at PI-RADS>=4. There was no statistical difference between pre- and post-CNN-review sensitivity and specificity for residents (McNemar test all p=1.00). Clinical radiologist specificity was significantly lower at PI-RADS>=3 compared to resident (p=0.002) but not CNN (p=0.07) assessment. Clinical radiologists detected all sPC at PI-RADS>=3 while CNN and residents missed 2 each, at an insignificant sensitivity difference (p=0.48). Radiologists missed only 3 sPC at PI-RADS>=4 while residents missed 7 sPC (p=0.07) and CNN missed 2 sPC (p=1.00).DISCUSSION
CNN demonstrates reasonable operating points in a prospective para-clinical assessment optimized by intermittent re-calibration using the sliding-windows technique. Overall, the nnUNet demonstrates comparable information extraction from images compared to both radiologists and residents over the entire ROC range. Operating points vary, with radiologists detecting more sPC at the given thresholds. Residents made more specific calls than suggested by CNN, at a specificity benefit at PI-RADS>=3 and >=4, however at the cost of lower sPC detection at the PI-RADS>=4 threshold.CONCLUSION
Fully-automated CNN detection of sPC confirms comparable performance to clinical and resident PI-RADS assessment in prospective case-by-case para-clinical assessment. The overall excellent performance of residents and limited change in case assessment after CNN review may reflect those residents quickly achieving a high level of prostate MRI interpretation skills and consequent confidence in their own assessment at our prostate MRI expert center. The tendency of higher specificity of resident reads likely reflects the lack of clinical consequences from research reads, allowing a more specific approach, while clinicians err on the side of not missing sPC, based on the experience built from frequent feedback from clinico-pathological image review conferences. Given the demonstrated high diagnostic capability of CNN, further study of the early prostate MRI learning curve with and without CNN use is motivated for the future.Acknowledgements
No acknowledgement found.References
1. Netzer N, Weißer C, Schelb P, et al. Fully Automatic Deep Learning in Bi-institutional Prostate Magnetic Resonance Imaging: Effects of Cohort Size and Heterogeneity. Investigative Radiology 2021;Publish Ahead of Print.
2. Schelb P, Kohl S, Radtke JP, et al. Classification of Cancer at Prostate MRI: Deep Learning versus Clinical PI-RADS Assessment. Radiology 2019;293(3):607-617.
3. Schelb P, Wang X, Radtke JP, et al. Simulated clinical deployment of fully automatic deep learning for clinical prostate MRI assessment. Eur Radiol 2021;31(1):302-313.
Figures
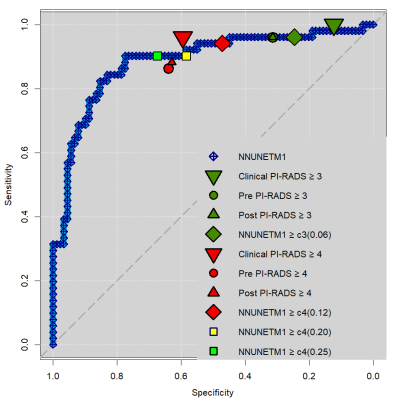
Figure 1: Receiver operating characteristics (ROC) curve of prospective CNN performance (blue diamonds). Superimposed are operating points (OP) of clinical radiologist assessment (large downward pointing triangles), para-clinical resident assessment before (circles) and after (small upward pointing triangles) review of CNN results, and CNN performance alone at calibrated thresholds (diamonds). OPs are shown for PI-RADS>=3 (green) and >=4 (red). Additional OPs are shown in light green and yellow (squares) at pre-set probability thresholds of 0.20 and 0.25 for comparison.