2832
Deep learning for prostate zonal segmentation robust to multicenter data1Electrical Engineering, Eindhoven University of Technology, Eindhoven, Netherlands, 2Radiology, Radboud university medical center, Nijmegen, Netherlands, 3Radiology, Netherlands Cancer Institute, Amsterdam, Netherlands
Synopsis
Prostate zonal segmentation is an important step for automated PCa diagnosis, MRI-guidedradiotherapy and focal treatment planning. Here we proposed a multi-channel U-Net for automatic prostate zonal segmentation, able to include multiple MRI sequences. Using a small, multicenter, multiparametric MRI dataset, we investigated its robustness towards the acquisition protocol and whether additional imaging sequences improve segmentation performance. Our results show that T2-weighted imaging alone is sufficient for successful prostate zonal segmentation. Despite using a small multicenter dataset, the models were robust towards the acquisition protocol and the performance was comparable to that obtained with larger datasets from a single institute.
Introduction
MRI prostate segmentation is an essential step for MRI-transrectral ultrasound fused guided biopsies and planning of MRI-guided prostate cancer (PCa) treatments that rely on daily treatment adaptation, such as (MRI-guided) radiotherapy. Zonal segmentation is useful for automated PCa diagnosis and focal treatment planning. However, manual segmentation is cumbersome and prone to interobserver variability1. Although convolutional neural networks (CNN) have shown great promise for object segmentation tasks in several fields, their application in medical imaging is often hampered by the need for large training datasets. Moreover, an optimal automated segmentation method should be invariant to data acquired using different scanners and protocols. In PCa, a multiparametric MRI imaging protocol is currently recommended, including T2-weighted (T2W), dynamic contrast-enhanced (DCE) imaging, and diffusion-weighted imaging; from the latter, maps of the apparent diffusion coefficient (ADC) are typically extracted. Up until now, mainly T2W imaging has been used for prostate (zonal) segmentation with only few studies comparing the performance between T2W-based and ADC-based segmentation2,3. In this study, we evaluated the robustness of CNN-based automatic prostate segmentation towards the acquisition protocol by training on a small, multicenter dataset and testing on a different unseen dataset from a different center. Using a multichannel strategy, we expanded the network to include also ADC and/or DCE images, and investigated whether the information provided by additional sequences improves the segmentation performance.Methods
The study included multiparametric MRI examinations from 75 PCa patients acquired in three institutions (Amsterdam UMC [AUMC], Netherlands Cancer Institute [NKI], and Radboudumc [RUMC]) as part of the Prostate Cancer Molecular Medicine study. Institute review board and ethical committee approval was obtained at each institution. Instutions used different scanners (Siemens and Philips), coils and field strengths, with and without endorectal coil. The whole prostate, central gland (CG), and peripheral zone (PZ) were delineated in consensus with a technical expert. A multichannel CNN was implemented based a 2D U-Net archicteture, with the number of channels determnined by the number of input imaging sequences. Base models were obtained by using as input T2W only, T2W+ADC, T2W+DCE, and T2W+ADC+DCE. For DCE, only the peak enhancement frame was used. To verify the model’s robustness towards the acquisition protocol, supervised domain adaptation (sDA) was performed. The four base models were first trained using data from 51 patients from two institutions (19 AUMC and 32 RUMC) using 5-fold cross validation; then sDA was performed by re-training with 16 patients, including 8 patients from the third institution (NKI). Both base models and sDA models were evaluated on the same set of 16 unseen NKI patients. Classification performance was assessed using the Dice similarity coefficient (DSC) and the Hausdorff distance (HD). Significant differences were investigated using Wilcoxon signed-rank test with a level of confidence of 0.01, adjusted for multiple comparison by the Dunn-Sìdak correction.Results
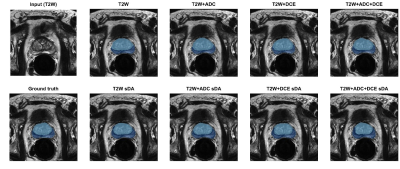
Although the base model using only T2W achieved slightly higher performance, results were generally comparable and no significant differences were found between all base models. Significant differences were also not found when comparing each base model with its corresponding sDA model. Figure 1 shows examples of the obtained predictions.Table 1 segmentation performance obtained on the test set. Results given as median and inter-quartile range in parenthesis.
| | Whole organ | PZ | CG | |||
| DSC | HD [mm] | DSC | HD [mm] | DSC | HD [mm] | |
| T2w | 0.93 (0.06) | 3.39 (0.79) | 0.86 (0.14) | 2.77 (1.04) | 0.82 (0.15) | 3.58 (0.88) |
| T2w sDA | 0.92 (0.07) | 3.49 (0.82) | 0.86 (0.12) | 2.88 (1.13) | 0.81 (0.15) | 3.67 (0.96) |
| T2w + ADC | 0.91 (0.08) | 3.39 (0.92) | 0.87 (0.13) | 2.77 (1.23) | 0.78 (0.15) | 3.84 (1.01) |
| T2w + ADC sDA | 0.91 (0.06) | 3.49 (1.01) | 0.86 (0.14) | 2.77 (1.23) | 0.80 (0.15) | 3.67 (1.11) |
| T2w + DCE | 0.91 (0.07) | 3.49 (0.93) | 0.86 (0.13) | 2.88 (1.23) | 0.80 (0.18) | 3.75 (1.18) |
| T2w + DCE sDA | 0.92 (0.07) | 3.49 (0.93) | 0.85 (0.14) | 2.88 (1.32) | 0.80 (0.17) | 3.75 (1.08) |
| T2w + ADC + DCE | 0.90 (0.08) | 3.49 (0.95) | 0.86 (0.14) | 2.88 (1.23) | 0.78 (0.18) | 3.75 (1.03) |
| T2w + ADC + DCE sDA | 0.91 (0.07) | 3.58 (0.90) | 0.85 (0.13) | 2.77 (1.23) | 0.78 (0.18) | 3.75 (1.08) |
Discussion
The performance results obtained in this study are comparable to those reported when using other deep learning approaches with larger training datasets3 and also to those of expert radiologists1. Our results suggest that T2w alone is capable to successfully segment the prostate. Additional imaging sequences, such as ADC or DCE, do not result in significant improvement. By using a small, multicenter dataset, with images acquired with different settings and protocols, our models showed to be robust and able to cope with new unseen datasets from a different institution, without the need for retraining. This is an encouraging finding as it suggests that by including sufficient variability in the training set, small datasets are sufficient to build a robust model that can be translated to a new institution without the need for further adaptations.Conclusions
Despite using a small multicenter dataset, the models were robust towards the acquisition protocol and the performance was comparable to that obtained with larger datasets from a single institute, indicating that training with heterogeneous data facilitates translatability to a new setting while maintaining performance.Acknowledgements
We acknowledge Chris Bangma for making the Prostate Cancer Molecular Medicine dataset available for this research.
References
1S. Montagne et al., “Challenge of prostate MRI segmentation on T2-weighted images: inter-observer variability and impact of prostate morphology.,” Insights Imaging, vol. 12, no. 1, p. 71, Jun. 2021, doi: 10.1186/s13244-021-01010-9.
2Zabihollahy, Fatemeh, et al. "Automated segmentation of prostate zonal anatomy on T2‐weighted (T2W) and apparent diffusion coefficient (ADC) map MR images using U‐Nets." Medical physics 46.7 (2019): 3078-3090.
3Cuocolo, Renato, et al. "Deep Learning Whole‐Gland and Zonal Prostate Segmentation on a Public MRI Dataset." Journal of Magnetic Resonance Imaging (2021).