2819
Towards a high-density packing white matter substrate generator for Monte-Carlo simulations
Juan Luis Villarreal Haro1, Remy Gardier1, Erick J Canales-Rodríguez1, Gabriel Girard1,2,3, Jean-Philippe Thiran1,2,3, and Jonathan Rafael-Patino1,3
1Signal Processing Laboratory (LTS5), Ecole Polytechnique Fédérale de Lausanne (EPFL), Lausanne, Switzerland, 2CIBM, Center for Biomedical Imaging, Lausanne, Switzerland, 3Radiology Department, Centre Hospitalier Universitaire Vaudois and University of Lausanne, Lausanne, Switzerland
1Signal Processing Laboratory (LTS5), Ecole Polytechnique Fédérale de Lausanne (EPFL), Lausanne, Switzerland, 2CIBM, Center for Biomedical Imaging, Lausanne, Switzerland, 3Radiology Department, Centre Hospitalier Universitaire Vaudois and University of Lausanne, Lausanne, Switzerland
Synopsis
One main goal of Diffusion-Weighted Magnetic Resonance Imaging is to infer microscopic tissue properties from the measured signal. The use of Monte-Carlo Simulations for DW-MRI on realistic substrates will help the study of DWI-MRI signals in controlled environments and to investigate, extract, and validate approaches for the understanding of white matter features. In this work, we present a novel framework for creating complex white matter phantoms with unprecedented characteristics like high packing density greater than 90% intra-axonal volume fraction and voxel size of (140 um)3.
Introduction
Monte-Carlo Simulation has emerged as a valuable tool for studying microstructure models in Diffusion-Weighted Magnetic Resonance Imaging (DW-MRI)[9]. They are particularly important when studying diffusion phenomena for complex scenarios with no analytical solutions, such as like axons with undulations, variable diameter, and local microdispersion, as well as the diffusion process in the extra cellular space. Monte-Carlo simulations, in contrast to standard methods, do not require any explicit model of the diffusion signal for a given geometry. Instead, a geometrical representation of the substrate in the form of a three-dimensional mesh is required. As a result, a realistic substrate will produce a DW-MRI signal containing relevant features. Previous works have addressed the problem of creating such realistic white matter substrates [3,4,8]. However, achieving realistic high packing densities is still an open challenge [7]. This work describes a new effort to create a novel and open-source numerical phantom generator for complex substrates. Notably, we show its ability to generate axonal packings with unprecedented intracellular volume fractions (ICVF) above 90% and a high sampling volume (number) big enough to represent the tissue properties of interest.Methods
We designed a novel substrate generator with parameterisable white matter features. In the first step, the prior distributions of the strands’ trajectories and radii is selected. Some examples of the priors selected are, parallel strands in a single fibre population and constant radius, single fibre populations with 5º of dispersion or two fiber populations crossing at 30º. The prior strand trajectories were taken from segmented data like [5], as is shown in Figure 2.In the second step, the strands are divided into a set of control spheres along their trajectories. Inspired by previous substrate generators [2,4], our generator optimizes a global cost function to jointly modify the size, radii and position of the control spheres of strands with desired features. Such features are constrained curvature, smooth transversal radii changes in strands, axon dispersion, spring force between control points, or non-overlapping strands (see Figure 1).
As a third step, we developed a post-processing framework based on the metaballs method (an inverse-square law for discretized spheres)[1]. Here, each strand is built in parallel using a local-sparse grid of the strand’s neighbourhood, which allows building the isosurface of the metaballed-strands with marching cubes algorithms. The fine-tuning of the hyperparameters of the post-processing allowed our generator to achieve high intra-cellular volume fractions on the prior strands’ distribution. Moreover, weighting the metaball thresholding individually allowed us to set grow-size constraints in the axon to maintain a similar prior radii distribution of the strands.
Results and Discussion
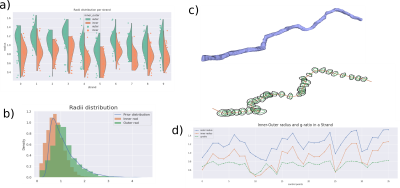
Our generator is able to create meshes for particularly large substrates in comparison to previous studies. For example, Figure 2, shows a substrate of size (140 μm)3 with an ICVF of 90%. Notably, the ICVF is higher than that reported in [3, 4, 8] where the highest values were in the range of 65-70%. Indeed, reported simulation substrates with dense axonal configurations only reached volumes around (35-100 μm)3 [3, 4, 8], whereas, in our preliminary results, we reached voxel size above (140 um)3. These highly-dense configurations, with high ICVF better mimic those from previous histological studies based on EM segmentation [10].Figure 3 shows a qualitative and quantitative shape analysis of the global and local radii distribution of the strands. The characteristics of the axonal shapes obtained in our framework are within range of interest to the WM axons obtained in analyses in Andersson et al. (2021) [5], made with high-resolution 3D synchrotron. Figure 4 shows an example of Monte-Carlo simulations made with the substrate from Figure 2. Simulations using complex substrates is essential to generate synthetic signals from different acquisition protocols to study the sensitivity of various acquisition protocols, and to characterize and validate existing models in more realistic scenarios.
The improvement in density packing, axonal shape, and voxel sizes are features that will lead to improved realism of Monte-Carlo simulations in DW-MRI.
Conclusion
The construction of brain virtual tissue for the generation of numerical phantoms is a crucial task for microstructure modelling. However, the complexity of the white matter axons configuration makes it a challenging task for today's methods, especially regarding generating realistic and densely packed substrates. In this work, we present a novel framework to generate realistic and ultra-packed virtual white matter. Our preliminary results show its potential to generate white matter configuration with unprecedented high packing densities above 90% intra-axonal volume fraction. Our results also show that the generated substrates have microstructural characteristics relevant for modelling the white matter microstructure [9]. Future work will focus on optimizing the substrate generator for large-scale substrates designed for connectivity estimation.Acknowledgements
This work is partly supported by the Swiss National Science Foundation under grant Nbr 205320_175974. We acknowledge access to the facilities and expertise of the CIBM Center for Biomedical Imaging, a Swiss research center of excellence founded and supported by Lausanne University Hospital (CHUV), University of Lausanne (UNIL), Ecole polytechnique fédérale de Lausanne (EPFL), University of Geneva (UNIGE) and Geneva University Hospitals (HUG). Erick J. Canales-Rodríguez was supported by the Swiss National Science Foundation (SNSF, Ambizione grant PZ00P2_185814).References
- Blinn, James F. "A generalization of algebraic surface drawing." ACM transactions on graphics (TOG) 1.3 (1982): 235-256.
- Close, Thomas G., et al. "A software tool to generate simulated white matter structures for the assessment of fibre-tracking algorithms." NeuroImage 47.4 (2009): 1288-1300.
- Callaghan, Ross, et al. "ConFiG: Contextual Fibre Growth to generate realistic axonal packing for diffusion MRI simulation." Neuroimage 220 (2020): 117107.
- Ginsburger, Kévin, et al. "MEDUSA: A GPU-based tool to create realistic phantoms of the brain microstructure using tiny spheres." NeuroImage 193 (2019): 10-24.
- Andersson, M., Kjer, H. M., Rafael-Patino, J., Pacureanu, A., Pakkenberg, B., Thiran, J. P., ... & Dyrby, T. B. (2020). Axon morphology is modulated by the local environment and impacts the noninvasive investigation of its structure–function relationship. Proceedings of the National Academy of Sciences, 117(52), 33649-33659.
- Lee, Hong-Hsi, Els Fieremans, and Dmitry S. Novikov. "Realistic Microstructure Simulator (RMS): Monte Carlo simulations of diffusion in three-dimensional cell segmentations of microscopy images." Journal of Neuroscience Methods 350 (2021): 109018.
- Rafael-Patino, Jonathan, et al. "Robust Monte-Carlo simulations in diffusion-MRI: Effect of the substrate complexity and parameter choice on the reproducibility of results." Frontiers in neuroinformatics 14 (2020): 8.
- Rafael-Patino, J., Girard, G., Truffet, R., Pizzolato, M., Thiran, J. P., & Caruyer, E. (2021, October). The Microstructural Features of the Diffusion-Simulated Connectivity (DiSCo) Dataset. In International Workshop on Computational Diffusion MRI (pp. 159-170). Springer, Cham.
- Novikov, Dmitry S., et al. "Quantifying brain microstructure with diffusion MRI: Theory and parameter estimation." NMR in Biomedicine 32.4 (2019): e3998.
- Liewald, Daniel, et al. "Distribution of axon diameters in cortical white matter: an electron-microscopic study on three human brains and a macaque." Biological cybernetics 108.5 (2014): 541-557.
Figures
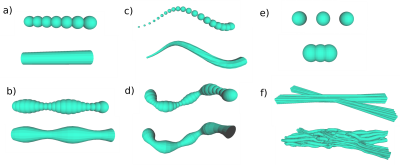
Generator’s substrate features. Panel a) shows the Control Spheres (CS) used to build strands b) Strand example built with a Cost Function (CF) for smooth radii changes between 2 consecutive CS c) CF for a constrained angle between 3 consecutive CS centroids; d) CF for local dispersion of strands; e) CF enforce Hooke’s springs law among 2 CS f) CF to disentangle overlappings of CS from different strands: before and after the optimization procedure with all CF.
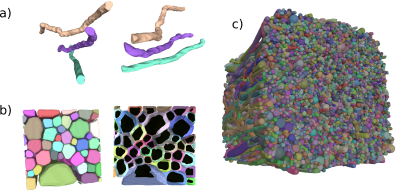
Results of the post-processing step. a) Three isolated strands from two points of view. The axons’ diameter and microstructural shape change non-symmetrically along the axons.. b) Packing substrate zoom-in showing the substrate’s compartments; the myelin wraps are coloured individually for each axon, and the dark inner mesh is the non-myelinated axon. c) Voxel example with dimension (140um)^3, 5º dispersion, prior gamma Γ(κ=2.2,θ =2) radii distribution, mean ratio of inner to outer radii (g-ratio) is of 0.7, and packing of 90% for the ICVF.
Particles’ trajectory of Monte-Carlo Brownian simulations in the mesh of the substrate of Figure 2. a,b) Trajectories in Intra-axonal space. The particles are inside the meshes of the de-myelinated axons.. The restricted motion shows the axonal shape. c,d) Trajectories in extra-axonal space. The particles are outside of the myelinated axons.. Hindered restricted motion of complex extracellular space in a voxel with 90% of ICVF.
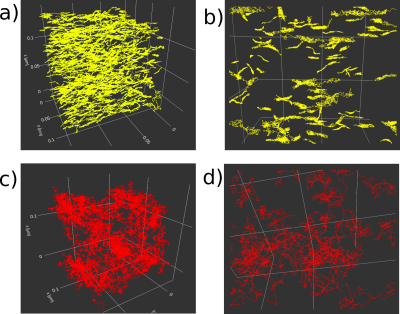
Particles’ trajectory of Monte-Carlo Brownian simulations in the mesh of Figure 2. Panel a) Trajectories in Intra-axonal space. The particles are inside the meshes of the de-myelinated axons. Panel b) Zoom-in. The restricted motion shows the axonal shape. Panel c) Trajectories in extra-axonal space. The particles are outside of the myelinated axons. Panel d) Zoom-in. Hindered restricted motion of complex extracellular space in a voxel with 90% of intracellular volume fraction.
DOI: https://doi.org/10.58530/2022/2819