2814
Cardiac motion modeling using DENSE imaging to study cardiac diffusion tensor MRI1CREATIS, Lyon, France, 2Department of Radiology, University Hospital Saint-Etienne, Saint Etienne, France, 3Department of Mechanical and Aerospace Engineering, University of Central Florida, Orlando, FL, United States
Synopsis
Cardiac Diffusion Tensor imaging (cDTI) is a powerful non-contrast technique that can characterize cellular structure in vivo. Despite motion-compensated diffusion encoding designs, cDTI remains sensitive to unwanted cardiac motion, particularly during diastole. The lack of knowledge of the detailed interaction between real cardiac motion and motion-compensated gradients prevents more advanced diffusion encoding designs. The objective of this work is to provide a new simulation framework to study the interaction between cardiac displacements estimated in vivo from DENSE imaging and motion-compensated diffusion encoding waveforms.
Introduction
Cardiac Diffusion Tensor imaging (cDTI) is a powerful non-contrast technique that probes the displacement of water molecules and from it infers the cellular structure in vivo. Advanced motion-compensated gradient designs have been proposed for cDTI1,2, such as first and second order nulled designs. These designs rely upon the assumption of linear velocity or linear acceleration of cardiac motion during the encoding of diffusion. These waveforms successfully compensate cardiac motion during most of systole, but generally not during diastole. No designs have been proposed to compensate cardiac motion in diastole, principally due to the lack of knowledge on the interaction between motion compensated designs and real cardiac displacements.Spatiotemporally resolved maps of cardiac displacements can be measured with DENSE3 MRI Although, from the acquired displacements, it is theoretically possible to estimate if the cardiac motion respects the assumption of linear velocity or acceleration, the complete interaction between cardiac motion, diffusion motion, and motion compensated waveforms is still unknown.
The objective of this work was to provide a new simulation framework to study the interaction between cardiac motion and motion-compensated diffusion encoding waveforms. This framework uses realistic cellular structure and a real cardiac displacement field measured in vivo using DENSE MRI to simulate motion-altered diffusion signal.
Methods
The simulation framework shown in the animated Figure 1 is composed of 4 steps:1) Cell structure generation: The cardiac structure was generated by inserting and connecting cardiomyocytes in a 0.5x0.5x0.5 mm3 voxel until a given ECV was reached. Cardiomyocytes are represented using cylinders 100µm long and 10-20µm in diameter. Subsequently, a look-up table (mask) for intra- and extra cellular compartments with resolution 1mm was generated.
2) Diffusion Displacement Simulation: The displacement of water molecules was simulated using a particle-based random-walk method with a native coefficient of diffusion D0 equal to 3x10-3mm2/s and 2.2x10-3mm2/s in the extra- and intra-cellular compartments4, respectively. These compartments were kept impermeable. A time step Dt=10µs was adopted and diffusion motion was linearly interpolated to 1µs in the MR simulations.
3) Cardiac Displacement Simulation: A 3D displacement field was applied to the voxel boundary (corners) and then linearly interpolated to the water molecules within the voxel to simulate the effect of cardiac deformation on the diffusion displacement distribution.
4) MR Simulation: Diffusion MR phase distribution were generated by numerically integrating the water molecules’ displacements and the diffusion encoding gradients with Dt=1ms.
First, nine cellular structures with ECV ranging from 0.15 to 1.0 and 25 repetitions using 10,000 water molecules each were simulated to validate the framework presented in Steps 1 & 2.
Subsequently, a single model using mean ECV=0.25 with N=1,000 particles (Steps 1-2) was used in each voxel of the mid short axis DENSE slice to simulate Steps 3 & 4. The cardiac displacement field was extracted from cine DENSE images acquired in a healthy volunteer on a 3T scanner (Prisma, Siemens). Six cine DENSE slices were acquired in short axis (SA) and long axis (LA) views during breath holds using 2D displacement encoding (ke=0.1 cycle/mm, balanced 3-point encoding, TE=1ms, TR=15ms, 2.5x2.5x8mm3). Unfiltered SA and LA 2D displacements were combined to generate a 3D displacement field. Monopolar (M0), first order (M0M1), and second order motion compensated (M0M1M2) waveforms were generated for a b-value of 350s/mm² resulting in a TE of 38, 54, 61ms and for twelve encoding directions.
For each step, the Mean Diffusivity (MD) and Fractional Anisotropy (FA) were extracted based on a tensor model and using only the water molecules in the extra-cellular compartment. MDwater, FAwater, first (E1water) and second (E2water) eigenvalues were calculated directly from the water displacement distribution. MDMR was calculated from the MR signal attenuation generated from the phase distribution. The deviations from the linear velocity (DLinVel) and linear acceleration (DLinAcc) hypotheses were computed for the acquired DENSE displacement field
Results
Figure 2 shows MDwater, FAwater, E1water and E2water reported as function of ECV. MDwater, E1water and E2water increase and FAwater decreases as ECV increases. All values converge to an isotropic water distribution for ECV=1. As E2water corresponds to water molecules that are originally more restricted, E2water increases more than E1water as ECV increases.Maps of MDwater generated after applying the cardiac motion on top of diffusion are shown in Figure 3. Due to motion corruption, MDwater significantly increases in all cardiac phases above the value without motion.
Mean diffusivity (MDMR) maps obtained after simulating the three (M0, M0M1, and M0M1M2) encoding diffusion waveforms are shown in Figure 4. M0M1 and M0M1M2 waveforms reduce the phase disruption due to cardiac deformation. Motion-compensated waveforms reduce MDMR across cardiac phases compared to M0 waveforms, but MDMR remains above the case without motion (Figure 5). DLinVel and DLinAcc are the smallest in mid-systole (200ms) and in diastasis (700ms), which are also local minima for MDMR.
Discussion and conclusion
In this work a new simulation framework enables studying the effect of real cardiac motion on the diffusion signal. Results indicate that cardiac motion contains high-order components that affect cardiac diffusion signal even when M0-M1-M2 motion-compensated gradients are adopted. This framework paves the way to design in silico new motion compensated waveforms to optimize cardiac diffusion imaging throughout the cardiac cycle.Acknowledgements
No acknowledgement found.References
1. Gamper U, Boesiger P, Kozerke S. Diffusion imaging of the in vivo heart using spin echoes–considerations on bulk motion sensitivity. Magn. Reson. Med. 2007;57:331–337 doi: 10.1002/mrm.21127.
2. Stoeck CT, Deuster C von, Genet M, Atkinson D, Kozerke S. Second-order motion-compensated spin echo diffusion tensor imaging of the human heart. Magn. Reson. Med. 2016;75:1669–1676 doi: 10.1002/mrm.25784.
3. Spottiswoode BS, Zhong X, Hess AT, et al. Tracking Myocardial Motion From Cine DENSE Images Using Spatiotemporal Phase Unwrapping and Temporal Fitting. IEEE Trans. Med. Imaging 2007;26:15–30 doi: 10.1109/TMI.2006.884215.
4. Rose JN, Nielles‐Vallespin S, Ferreira PF, Firmin DN, Scott AD, Doorly DJ. Novel insights into in‐vivo diffusion tensor cardiovascular magnetic resonance using computational modeling and a histology‐based virtual microstructure. Magn. Reson. Med. 2019;81:2759–2773 doi: 10.1002/mrm.27561.
Figures
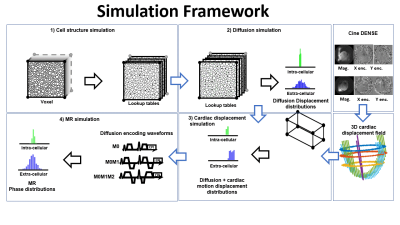
Figure 1: The simulation framework is composed of 4 steps: 1) Cell structures are generated and a look-up table is built. 2) The diffusion displacements of water molecules are simulated in the look-up table (mask). 3) Cardiac displacements are extracted from DENSE images and applied to deform the voxel containing the water molecules. 4) the MR signal is generated by numerical integration of the diffusion displacements and the diffusion encoding waveforms.
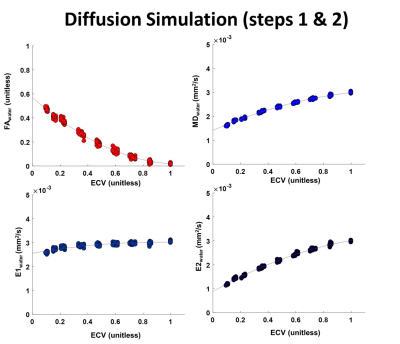
Figure 2: Fraction of Anisotropy (FA), Mean diffusivity (MD), First (E1) and second (E2) eigenvalues as a function of Extra-cellular volume (ECV). Values were calculated from the displacement of the water molecules in the extracellular space only. 9 structures spanning a range of ECV ratio from 0.15 to 1 were simulated and repeated 25 times.
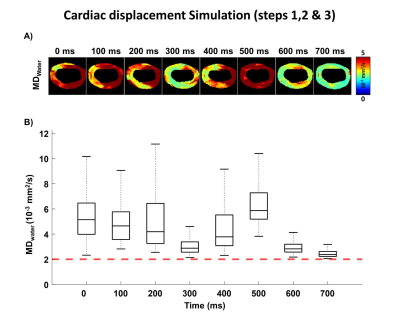
Figure 3: A) Mean Diffusivity (MDwater) maps generated after applying the cardiac displacement field. B) Corresponding MDwater distribution at each cardiac phase. The red dashed line represent the MD obtained without cardiac motion (MD=2 .10-3 mm2/s). MDwater is significantly higher than 2 .10-3 mm2/s due to motion corruption.
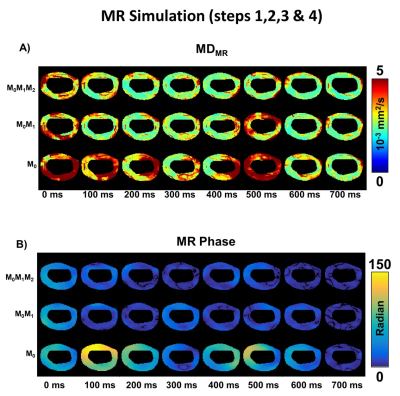
Figure 4: A) Mean Diffusivity (MDMR) maps at each cardiac phase are generated after simulating the MR signal for Monopolar (M0), first order (M0M1), and second order motion compensated (M0M1M2) waveforms. B) Corresponding phase of the MR signal for the M0, M0M1, and M0M1M2 waveforms.
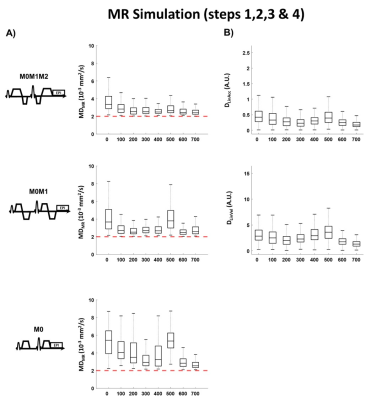
Figure 5: A) Distribution of Mean Diffusivity (MDMR) during the cardiac cycle generated after simulating the MR signal for Monopolar (M0), first order (M0M1), and second order motion compensated (M0M1M2) waveforms. B) Deviation from linear velocity DLinVel and linear acceleration DLinAcc calculated for the cardiac displacement field acquired with cine DENSE imaging.